Does davidad’s uploading moonshot work?
davidad has a 10-min talk out on a proposal about which he says: “the first time I’ve seen a concrete plan that might work to get human uploads before 2040, maybe even faster, given unlimited funding”.
I think the talk is a good watch, but the dialogue below is pretty readable even if you haven’t seen it. I’m also putting some summary notes from the talk in the Appendix of this dialoge.
I think of the promise of the talk as follows. It might seem that to make the future go well, we have to either make general AI progress slower, or make alignment progress differentially faster. However, uploading seems to offer a third way: instead of making alignment researchers more productive, we “simply” run them faster. This seems similar to OpenAI’s Superalignment proposal of building an automated alignment scientist—with the key exception that we might plausibly think a human-originated upload would have a better alignment guarantee than, say, an LLM.
I decided to organise a dialogue on this proposal because it strikes me as “huge if true”, yet when I’ve discussed this with some folks I’ve found that people generally don’t seem that aware of it, and sometimes raise questions/confusions/objections that neither me nor the talk could answer.
I also invited Anders Sandberg, who co-authored the 2008 Whole Brain Emulation roadmap with Nick Bostrom, and co-organised the Foresight workshop on the topic where Davidad presented his plan, and Lisa Thiergart from MIRI, who also gave a talk at the workshop and has previously written about whole brain emulation here on LessWrong.
The dialogue ended up covering a lot of threads at the same time. I split them into six separate sections, that are fairly readable independently.
Barcoding as blocker (and synchrotrons)
One important thing is “what are the reasons that this uploading plan might fail, if people actually tried it.” I think the main blocker is barcoding the transmembrane proteins. Let me first lay out the standard argument for why we “don’t need” most of the material in the brain, such as DNA methylation, genetic regulatory networks, microtubules, etc.
Cognitive reaction times are fast.
That means individual neuron response times need to be even faster.
Too fast for chemical diffusion to go very far—only crossing the tiny synaptic gap distance.
So everything that isn’t synaptic must be electrical.
Electricity works in terms of potential differences (voltages), and the only parts of tissue where you can have a voltage difference is across a membrane.
Therefore, all the cognitive information processing supervenes on the processes in the membranes and synapses.
Now, people often make a highly unwarranted leap here, which is to say that all we need to know is the geometric shapes and connection graph of the membranes and synapses. But membranes and synapses are not made of homogenous membraneium and synapseium. Membranes are mostly made of phospholipid bilayers, but a lot of the important work is done by trans-membrane proteins: it is these molecules that translate incoming neurotransmitters into electrical signals, modulate the electrical signals into action potentials, cause action potentials to decay, and release the neurotransmitters at the axon terminals. And there are many, many different trans-membrane proteins in the human nervous system. Most of them probably don’t matter or are similar enough that they can be conflated, but I bet there are at least dozens of very different kinds of trans-membrane proteins (inhibitory vs excitatory, different time constants, different sensitivity to different ions) that result in different information-processing behaviours. So we need to not just see the neurons and synapses, but the quantitative densities of each of many different kinds of transmembrane proteins throughout the neuron cell membranes (including but not limited to at the synapse).
Sebastian Seung famously has a hypothesis in his book about connectomics that there are only a few hundred distinct cell types in the human brain that have roughly equivalent behaviours at every synapse, and that we can figure out which type each cell is just from the geometry. But I think this is unlikely given the way that learning works via synaptic plasticity: i.e. a lot of what is learned, at least in memory (as opposed to skills) is represented by the relative receptor densities at synapses. However, it may be that we are lucky and purely the size of the synapse is enough. In this case actually we are extremely lucky because the synchrotron solution is far more viable, and that’s much faster than expansion microscopy. With expansion microscopy, it seems plausible to tag a large number of receptors by barcoding fluorophores in a similar way to how folks are now starting to barcode cells with different combinations of fluorophores (which solves an entirely different problem, namely redundancy for axon tracing). However, the most likely way for the expansion-microscopy-based plan to fail is that we cannot use fluorescence to barcode enough transmembrane proteins.
What options are there for overcoming this, and how tractable are they?
For barcoding, there are only a couple different technical avenues I’ve thought of so far. To some extent they can be combined.
“serial” barcoding, in which we find a protocol for washing out a set of tags in an already-expanded sample, and then diffusing a new set of tags, and being able to repeat this over and over without the sample degrading too much from all the washing.
“parallel” barcoding, in which we conjugate several fluorophores together to make a “multicolored” fluorophore (literally like a barcode in the visible-light spectrum). This is the basic idea that is used for barcoding cells, but the chemistry is very different because in cells you can just have different concentrations of separate fluorophore molecules floating around, whereas receptors are way too small for that and you need to have one of each type of fluorophore all kind of stuck together as one molecule. Chemistry is my weakness, so I’m not very sure how plausible this is, but from a first-principles perspective it seems like it might work.
the synchrotron solution is far more viable
On synchotrons / particle accelerator x-rays: Logan Collins estimates they would take ~1 yr of sequential effort for a whole human brain (which thus also means you could do something like a mouse brain in a few hours, or an organoid in minutes, for prototyping purposes). But I’m confused why you suggest that as an option that’s differentially compatible with only needing lower resolution synaptic info like size.
Could you not do expansion microscopy + synchrotron? And if you need the barcoding to get what you want, wouldn’t you need it with or without synchrotron?
So, synchrotron imaging uses X-rays, whereas expansion microscopy typically uses ~visible light fluorescence (or a bit into IR or UV). (It is possible to do expansion and then use a synchrotron, and the best current synchrotron pathway does do that, but the speed advantages are due to X-rays being able to penetrate deeply and facilitate tomography.) There are a lot of different indicator molecules that resonate at different wavelengths of ~visible light, but not a lot of different indicator atoms that resonate at different wavelengths of X-rays. And the synchrotron is monochromatic anyway, and its contrast is by transmission rather than stimulated emission. So for all those reasons, with a synchrotron, it’s really pushing the limits to get even a small number of distinct tags for different targets, let alone spectral barcoding. That’s the main tradeoff with synchrotron’s incredible potential speed.
One nice thing with working in visible light rather than synchrotron radiation is that the energies are lower, and hence there is less disruption of the molecules and structure.
Andreas Schaefer seems to be making great progress with this, see e.g. here. I have updated downward in conversations with him that sample destruction will be a blocker.
Also, there are many modalities that have been developed in visible light wavelengths that are well understood.
This is a bigger concern for me, especially in regards to spectral barcodes.
End-to-end iteration as blocker (and organoids, holistic processes, and exotica)
I think the barcoding is a likely blocker. But I think the most plausible blocker is a more diffuse problem: we do not manage to close the loop between actual, running biology, and the scanning/simulation modalities so that we can do experiments, adjust simulations to fit data, and then go back and do further experiments—including developing new scanning modalities. My worry here is that while we have smart people who are great at solving well-defined problems, the problem of setting up a research pipeline that is good at iterating at generating well-defined problems might be less well-defined… and we do not have a brilliant track record of solving such vague problems.
Yeah, this is also something that I’m concerned that people who take on this project might fail to do by default, but I think we do have the tech to do it now: human brain organoids. We can manufacture organoids that have genetically-human neurons, at a small enough size where the entire thing can be imaged dynamically (i.e. non-dead, while the neurons are firing, actually collecting the traces of voltage and/or calcium), and then we can slice and dice the organoid and image it with whatever static scanning modality, and see if we can reproduce the actual activity patterns based on data from the static scan. This could become a quite high-throughput pipeline for testing many aspects of the plan (system identification, computer vision, scanning microscopes, sample prep/barcoding, etc.).
What is the state of the art of “uploading an organoid”?
(I guess one issue is that we don’t have any “validation test”: we just have neural patterns, but the organoid as a whole isn’t really doing any function. Whereas, for example, if we tried uploading a worm we could test whether it remembers foraging patterns learnt pre-upload)
I’m not sure if I would know about it, but I just did a quick search and found things like “Functional neuronal circuitry and oscillatory dynamics in human brain organoids” and “Stretchable mesh microelectronics for biointegration and stimulation of human neural organoids”, but nobody is really trying to “upload an organoid”. They are already viewing the organoids as more like “emulations” on which one can do experiments in place of human brains; it is an unusual perspective to treat an organoid as more like an organism which one might try to emulate.
On the general theme of organoids & shortcuts:
I’m thinking about what are key blockers in dynamic scanning as well later static slicing & imaging. I’m probably not fully up to date on the state of the art of organoids & genetic engineering on them. However, if we are working on the level of organoids, couldn’t we plausibly directly genetically engineer (or fabrication engineer) them to make our life easier?
ex. making the organoid skin transparent (visually or electrically)
ex. directly have the immunohistochemistry applied as the organoid grows / is produced
ex. genetically engineer them such that the synaptic components we are most interested in are fluorescent
...probably there are further such hacks that might speed up the “Physically building stuff and running experiments” rate-limit on progress
(ofc getting these techniques to work will also take time, but at appropriate scales it might be worth it)
We definitely can engineer organoids to make experiments easier. They can simply be thin enough to be optically transparent, and of course it’s easier to grow them without a skull, which is a plus.
In principle, we could also make the static scanning easier, but I’m a little suspicious of that, because in some sense the purpose of organoids in this project would be to serve as “test vectors” for the static scanning procedures that we would want to apply to non-genetically-modified human brains. Maybe it would help to get things off the ground to help along the static scanning with some genetic modification, but it might be more trouble than it’s worth.
Lisa, here’s a handwavy question. Do you have an estimate, or the components of an estimate, for “how quickly could we get a ‘Chris Olah-style organoid’—that is, the smallest organoid that 1) we fully understood, in the relevant sense and also 2) told us at least something interesting on the way to whole brain emulation?”
(Also, I don’t mean that it would be an organoid of poor Chris! Rather, my impression is that his approach to interpretability, is “start with the smallest and simplest possible toy system that you do not understand, then understand that really well, and then increase complexity”. This would be adapting that same methodology)
Wow that’s a fun one, and I don’t quite know where to start. I’ll briefly read up on organoids a bit. [...reading time...] Okay so after a bit of reading, I don’t think I can give a great estimate but here are some thoughts:
First off, I’m not sure how much the ‘start with smallest part and then work up to the whole’ works well for brains. Possibly, but I think there might be critical whole brain processes we are missing (that change the picture a lot). Similarly, I’m not sure how much starting with a simpler example (say a different animal with a smaller brain) will reliably transfer, but at least some of it might. For developing the needed technologies it definitely seems helpful.
That said, starting small is always easier. If I were trying to come up with components for an estimate I’d consider:
looking at non-human organoids
comparisons to how long similar efforts have been taking: C. elegans is a simpler worm organism some groups have worked on trying to whole brain upload, which seems to be taking longer than experts predicted (as far as I know the efforts have been ongoing for >10 years and not yet concluded, but there has been some progress). Though, I think this is affected for sure by there not being very high investment in these projects and few people working on it.
trying to define what ‘fully understood’ means: perhaps being able to get within x% error on electrical activity prediction and/or behavioural predictions (if the organism exhibits behaviors)
There’s definitely tons more to consider here, but I don’t think it makes sense for me to try to generate it.
I’m not sure how much the ‘start with smallest part and then work up to the whole works’ well for brains. Possibly, but I think there might be critical whole brain processes we are missing (that change the picture a lot).
One simple example of a “holistic” property that might prove troublesome is oscillatory behavior, where you need a sufficient number of units linked in the right way to get the right kind of oscillation. The fun part is that you get oscillations almost automatically from any neural system with feedback, so distinguishing merely natural frequency oscillations (e.g. the gamma rhythm seems to be due to fast inhibitory interneurons if I remember right) and the functionally important oscillations (if any!) will be tricky. Merely seeing oscillations is not enough, we need some behavioral measures.
There is likely a dynamical balance between microstructure-understanding based “IKEA manual building” of the system and the macrostructure-understanding “carpentry” approach. Setting the overall project pipeline in motion requires having a good adaptivity on this.
Wouldn’t it be possible to bound the potentially relevant “whole brain processes”, by appealing to a version of Davidad’s argument above that we can neglect a lot of modalities and stuff, because they operate at a slower timescale than human cognition (as verified for example by simple introspection)?
I think glia are also involved in modulating synaptic transmissions as well as the electric conductivity of a neuron (definitely the speed of conductance), so I’m not sure the speed of cognition argument necessarily disqualifies them and other non-synaptic components as relevant to an accurate model. This paper presents the case of glia affecting synaptic plasticity and being electrically active. Though I do think that argument seems to be valid for many components, which clearly cannot effect the electrical processes at the needed time scales.
With regards to “whole brain processes” what I’m gesturing at is there might be top level control or other processes running without whose inputs the subareas’ function cannot be accurately observed. We’d need to have an alternative way of inputting the right things into the slice or subarea sample to generate the type of activity that actually occurs in the brain. Though, it seems we can focus on the electrical components of such a top level process in which case I wouldn’t see an obvious conflict between the two arguments. I might even expect the electrical argument to hold more, because of the need to travel quickly between different (far apart) brain areas.
Yeah, sorry, I came across as endorsing a modification of the standard argument that rules out too many aspects of brain processes as cognition-relevant, where I only rule back in neural membrane proteins. I’m quite confident that neural membrane proteins will be a blocker (~70%) whereas I am less confident about glia (~30%) but it’s still very plausible that we need to learn something about glial dynamics in order to get a functioning human mind. However, whatever computation the glia are doing is also probably based on their own membrane proteins! And the glia are included in the volume that we have to scan anyway.
Yeah, makes sense. I also am thinking that maybe there’s a way to abstract out the glia by considering their inputs as represented in the membrane proteins. However, I’m not sure whether it’s cheaper/faster to represent the glia vs. needing to be very accurate with the membrane proteins (as well as how much of a stretch it is to assume they can be fully represented there).
My take on glia is that they may represent a sizeable pool of nonlinear computation, but it seems to run slowly compared to the active spikes of neurons. That may require lots of memory, but less compute. But real headache is that there is relatively little research on glia (despite their very loyal loyalists!) compared to those glamorous neurons. Maybe they do represent a good early target for trying to define a research subgoal of characterizing them as completely as possible.
Overall, the “exotica” issue is always annoying—there could be an infinite number of weird modalities we are missing, or strange interactions requiring a clever insight. However, I think there is a pincer maneouver here where we try to constrain it by generating simulations from known neurophysiology (and nearby variations), and perhaps by designing experiments to estimate degrees of unknown degrees of freedom (much harder, but valuable). As far as I know the latter approach is still not very common, my instant association is to how statistical mechanics can estimate degrees of freedom sensitively from macroscopic properties (leading, for example, to primordial nucleogenesis to constrain elementary particle physics to a surprising degree). This is where I think a separate workshop/brainstorm/research minipipeline may be valuable for strategizing.
Using AI to speed up uploading research
As AI develops over the coming years, it will speed up some kinds of research and engineering. I’m wondering: for this proposal, which parts could future AI accelerate? And perhaps more interestingly: which parts would not be easily accelerable (implying that work on those parts sooner is more important)?
The frame I would use is to sort things more like along a timeline of when AI might be able to accelerate them, rather than a binary easy/hard distinction. (Ultimately, a friendly superintelligence would accelerate the entire thing—which is what many folks believe friendly superintelligence ought to be used for in the first place!)
The easiest parts to accelerate are the computer vision. In terms of timelines, this is in the rear-view mirror, with deep learning starting to substantially accelerate the processing of raw images into connectomic data in 2017.
The next easiest is the modelling of the dynamics of a nervous system based on connectomic data. This is a mathematical modelling challenge, which needs a bit more structure than deep learning traditionally offers, but it’s not that far off (e.g. with multimodal hypergraph transformers).
The next easiest is probably more ambitious microscopy along the lines of “computational photography” to extract more data with fewer electrons or photons by directing the beams and lenses according to some approximation of optimal Bayesian experimental design. This has the effect of accelerating things by making the imaging go faster or with less hardware.
The next easiest is the engineering of the microscopes and related systems (like automated sample preparation and slicing). These are electro-optical-mechanical engineering problems, so will be harder to automate than the more domain-restricted problems above.
The hardest to automate is the planning and cost-optimization of the manufacturing of the microscopes, and the management of failures and repairs and replacements of parts, etc. Of course this is still possible to automate, but it requires capabilities that are quite close to the kind of superintelligence that can maintain a robot fleet without human intervention.
The hardest to automate is the planning and cost-optimization of the manufacturing
An interesting related question is “if you had an artificial superintelligence suddenly appear today, what would be its ‘manufacturing overhang’? How long it would it take it to build the prerequisite capacity, starting with current tools?”
Could AI assistants help us build a research pipeline? I think so. Here is a sketch: I run a bio experiment, I scan the bio system, I simulate it, I get data that disagrees. Where is the problem? Currently I would spend a lot of effort trying to check my simulator for mistakes or errors, then move on to check whether the scan data was bad, then if the bio data was bad, and then if nothing else works, positing that maybe I am missing a modality. Now, with good AI assistants I might be able to (1) speed up each of these steps, including using multiple assistants running elaborate test suites. (2) automate and parallelize the checking. But the final issue remains tricky: what am I missing, if I am missing something? This is where we have human level (or beyond) intelligence questions, requiring rather deep understanding of the field and what is plausible in the world, as well as high level decisions on how to pursue research to test what new modalities are needed and how to scan for them. Again, good agents will make this easier, but it is still tricky work.
What I suspect could happen for this to fail is that we run into endless parameter-fiddling, optimization that might hide bad models by really good fits of floppy biological systems, and no clear direction for what to research to resolve the question. I worry that it is a fairly natural failure mode, especially if the basic project produces enormous amount of data that can be interpreted very differently. Statistically speaking, we want identifiability. But not just of the fit to our models, but to our explanations. And this is where non-AGI agents have not yet demonstrated utility.
Yeah, so, overall, it seems “physically building stuff”, “running experiments” and “figure out what you’re missing” are some of where the main rate-limiters lie. These are the hardest-to-automate steps of the iteration loop, that prevent an end-to-end AI assistant helping us through the pipeline.
Firstly though, it seems important to me how frequently you run into the rate limiter. If “figuring out what you’re missing” is something you do a few times a week, you could still be sped up a lot by automating the other parts of the pipeline until you can run this problem a few times per day.
But secondly I’m interested in answers digging one layer down of concreteness—which part of the building stuff is hard, and which part of the experiment running is hard? For example: Anders had the idea of “the neural pretty printer”: create ground truth artificial neural networks performing a known computation >> convert them into a Hodgkin-Huxley model (or similar) >> fold that up into a 3D connectome model >> simulate the process of scanning data from that model—and then attempt to reverse engineer the whole thing. This would basically be a simulation pipeline for validating scanning set-ups.
To the extent that such simulation is possible, those particular experiments would probably not be the rate-limiting ones.
The neural pretty printer is an example of building a test pipeline: we take a known simulatable neural system, convert it into a plausible biological garb and make fake scans of it according to the modalities we have, and then try to get our interpretation methods reconstruct it. This is great, but eventually limited. The real research pipeline will have to contain (and generate) such mini-pipelines to ensure testability. There is likely an organoid counterpart. Both have a problem of making systems to test the scanning based on somewhat incomplete (or really incomplete!) data.
Training frontier models to predict neural activity instead of next token
Lisa, you said in our opening question brainstorm:
1. Are there any possible shortcuts to consider? If there are, that seems to make this proposal even more feasible.
1a. Maybe something like, I can imagine there are large and functional structural similarities across different brain areas. If we can get an AI or other generative system to ‘fill in the gaps’ of more sparse tissue samples, and test whether the reconstructed representation is statistically indistinguishable from the dynamic data collected [(with the aim of figuring out the statics-to-dynamics map)] then we might be able to figure out what density of tissue sampling we need for full predictability. (seems plausible that we don’t need 100% tissue coverage, especially in some areas of the brain?). Note, it also seems plausible to me that one might be missing something important that could show up in a way that wasn’t picked up in the dynamic data, though that seems contingent on the quality of the dynamic data.
1b. Given large amounts of sparsity in neural coding, I wonder if there are some shortcuts around that too. (Granted though that the way the sparsity occurs seems very critical!)
Somewhat tangentially, this makes me wonder about taking this idea to its limit: what about the idea of “just” training a giant transformer to, instead of predicting next tokens in natural language, predicting neural activity? (at whatever level of abstractions is most suitable.) “Imitative neural learning”. I wonder if that would be within reach of the size of models people are gearing up to train, and whether it would better preserve alignment guarantees.
Hmm, intuitively this seems not good. On the surface level, two reasons come to mind:
1. I suspect the element of “more reliably human-aligned” breaks or at least we have less strong reasons to believe this would be the case than if the total system also operates on the same hardware structure (so to say, it’s not actually going to be run on something carbon-based). Though I’m also seeing the remaining issue of: “if it looks like a duck (structure) and quacks like a duck (behavior), does that mean it’s a duck?”. At least we have the benefit of deep structural insight as well as many living humans as a prediction on how aligned-ness of these systems turns out in practice. (That argument holds in proportion to how faithful of an emulation we achieve.)
2. I would be really deeply interested in whether this would work. It is a bit reminiscent of the Manifund proposal Davidad and I have open currently, where the idea is to see if human brain data can improve performance on next token prediction and make it more human preference aligned. At the same time, for the ‘imitative neural learning’ you suggest (btw I suspect it would be feasible with GPT4/5 levels, but I see the bottleneck/blocker in being able to get enough high quality dynamic brain data) I think I’d be pretty worried that this would turn into some dangerous system (which is powerful, but not necessarily aligned).
2/a Side thought: I wonder how much such a system would in fact be constrained to human thought processes (or whether it would gradient descend into something that looks input and output similar, but in fact is something different, and behaves unpredictably in unseen situations). Classic Deception argument I guess (though in this case without an implication of some kind of intentionality on the system’s side, just that it so happens bc of the training process and data it had)
I basically agree with Lisa’s previous points. Training a transformer to imitate neural activity is a little bit better than training it to imitate words, because one gets more signal about the “generators” of the underlying information-processing, but misgeneralizing out-of-distribution is still a big possibility. There’s something qualitatively different that happens if you can collect data that is logically upstream of the entire physical information processing conducted by the nervous system — you can then make predictions about that information-processing deductively rather than inductively (in the sense of the problem of induction), so that whatever inductive biases (aka priors) are present in the learning-enabled components end up having no impact.
“Human alignment”: one of the nice things with human minds is that we understand roughly how they work (or at least the signs that something is seriously wrong). Even when they are not doing what they are supposed to do, the failure modes are usually human, all too human. The crux here is whether we should expect to get human-like systems, or “humanish” systems that look and behave similar but actually work differently. The structural constraints from Whole Brain Emulation are a good reason to think more of the former, but I suspect many still worry about the latter because maybe there are fundamental differences in simulation from reality. I think this will resolve itself fairly straightforwardly since—by assumption if this project gets anywhere close to succeeding—we can do a fair bit of experimentation on whether the causal reasons for various responses look like normal causal reasons in the bio system. My guess is that here Dennett’s principle that the simplest way of faking many X is to be X. But I also suspect a few more years of practice with telling when LLMs are faking rather than grokking knowledge and cognitive steps will be very useful and perhaps essential for developing the right kind of test suite.
How to avoid having to spend $100B and and build 100,000 light-sheet microscopes
“15 years and ~$500B” ain’t an easy sell. If we wanted this to be doable faster (say, <5 years), or cheaper (say, <$10B): what would have to be true? What problems would need solving? Before we finish, I am quite keen to poke around the solution landscape here, and making associated fermis and tweaks.
So, definitely the most likely way that things could go faster is that it turns out all the receptor densities are predictable from morphology (e.g. synaptic size and “cell type” as determined by cell shape and location within a brain atlas). Then we can go ahead with synchrotron (starting with organoids!) and try to develop a pipeline that infers the dynamical system from that structural data. And synchrotron is much faster than expansion.
it turns out all the receptor densities are predictable from morphology
What’s the fastest way you can see to validating or falsifying this? Do you have any concrete experiments in mind that you’d wish to see run if you had a magic wand?
Unfortunately, I think we need to actually see the receptor densities in order to test this proposition. So transmembrane protein barcoding still seems to me to be on the critical path in terms of the tech tree. But if this proposition turns out to be true, then you won’t need to use slow expansion microscopy when you’re actually ready to scan an entire human brain—you only need to use expansion microscopy on some samples from every brain area, in order to learn a kind of “Rosetta stone” from morphology to transmembrane protein(/receptor) densities for each cell type.
So, for the benefit of future readers (and myself!) I kind of would like to see you multiply 5 numbers together to get the output $100B (or whatever your best cost-esimate is), and then do the same thing in the fortunate synchrotron world, to get a fermi of how much things would cost in that world.
Ok. I actually haven’t done this myself before. Here goes.
The human central nervous system has a volume of about 1400 cm^3.
When we expand it for expansion microscopy, it expands by a factor of 11 in each dimension, so that’s about 1.9 m^3. (Of course, we would slice and dice before expanding...)
Light-sheet microscopes can cover a volume of about 10^4 micron^3 per second, which is about 10^-14 m^3 per second.
That means we need about 1.9e14 microscope-seconds of imaging.
If the deadline is 10 years, that’s about 3e8 seconds, so we need to build about 6e5 microscopes.
Each microscope costs about $200k, so 6e5 * $200k is $120B. (That’s not counting all the R&D and operations surrounding the project, but building the vast array of microscopes is the expensive part.)
Pleased by how close that came out to the number I’d apparently been citing before. (Credit is due to Rob McIntyre and Michael Andregg for that number, I think.)
Similarly, looking at Logan’s slide:
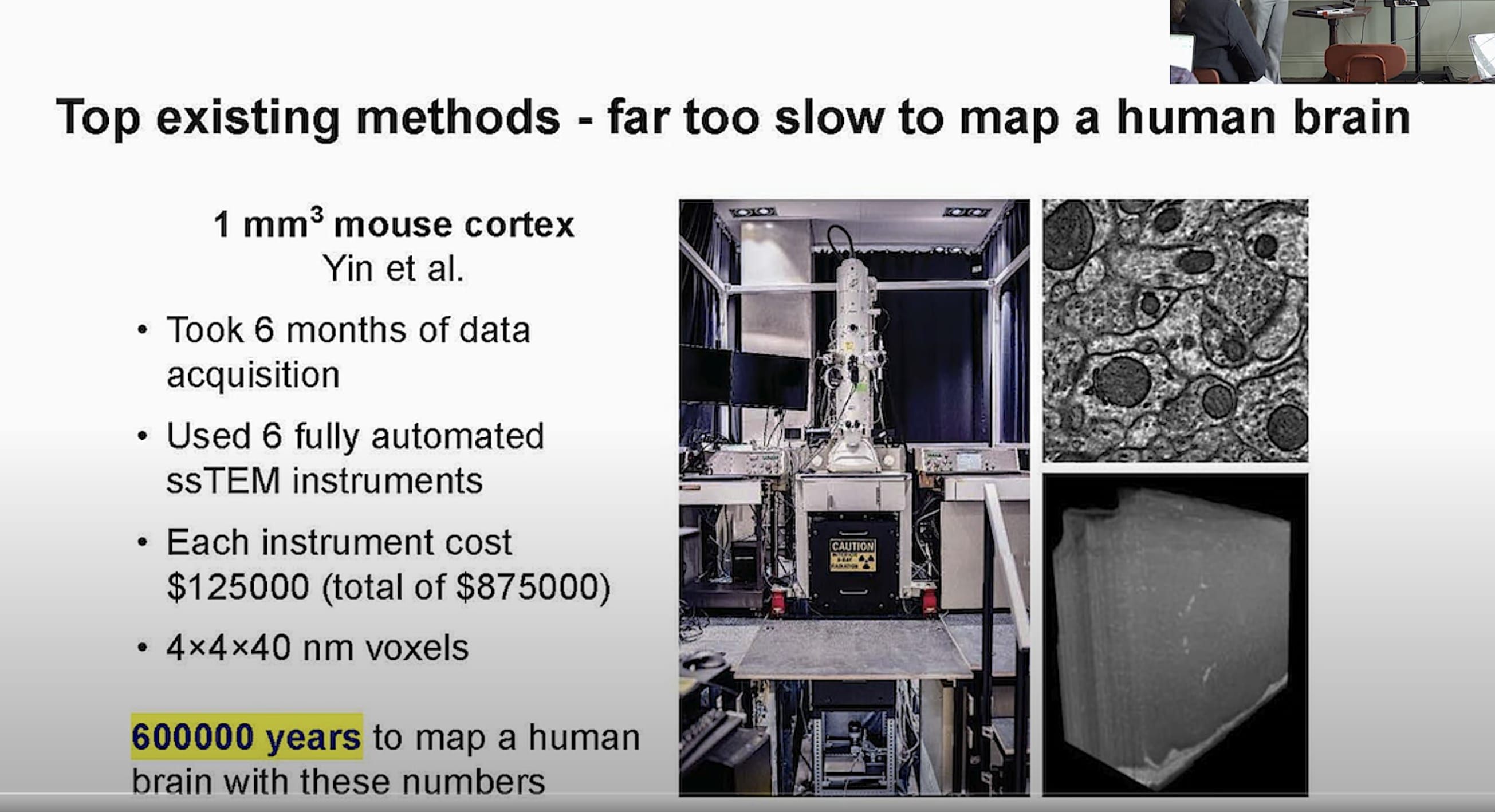
$1M for instruments, and needing 600k years. So, make 100k microscopes and run them in parallel for 6 years and then you get $100B…
That system is a bit faster than ExM, but it’s transmission electron microscopy, so you get roughly the same kind of data as synchrotron anyway (higher resolution, but probably no barcoded receptors)
Now for the synchrotron cost estimate.
Synchrotron imaging has about 300nm voxel size, so to get accurate synapse sizes we would still need to do expansion to 1.9 m^3 of volume.
Synchrotron imaging has a speed of about 600 s/mm^3, but it seems uncontroversial that this may be improved by an order of magnitude with further R&D investment.
That multiplies to about 3000 synchrotron-years.
To finish in 10 years, we would need to build 300 synchrotron beamlines.
Each synchrotron beamline costs about $10M.
That’s $3B in imaging infrastructure. A bargain!
That’s very different from this estimate. Thoughts?
One difference off the bat − 75nm voxel size is maybe enough to get a rough connectome, but not enough to get precise estimates of synapse size. I think we’d need to go for 11x expansion. So that’s about 1 order of magnitude, but there’s still 2.5 more to account for. My guess is that this estimate is optimistic about combining multiple potential avenues to improve synchrotron performance. I do see some claims in the literature that more like 100x improvement over the current state of the art seems feasible.
What truly costs money in projects? Generally, it is salaries and running costs times time, plus the instrument/facilities costs as a one-time cost. One key assumption in this moonshot is that there is a lot of scalability so that once the basic setup has been made it can be replicated ever cheaper (Wrightean learning, or just plain economies of scale). The faster the project runs, the lower the first factor, but the uncertainty about whether all relevant modalities have been covered will be greater. There might be a rational balance point between rushing in and likely having to redo a lot, and being slow and careful but hence getting a lot of running costs. However, from the start the difficulty is very uncertain, making the actual strategy (and hence cost) plausibly a mixture model.
The path to the 600,000 microscopes is of course to start with 6, doing the testing and system integration while generating the first data for the initial mini-pipelines for small test systems. As that proves itself one can scale up to 60 microscopes for bigger test systems and smaller brains. And then 600, 6,000 and so on.
Hundreds of thousands of microscopes seem to me like an… issue. I’m curious if you have something like a “shopping list” of advances or speculative technologies that could bring that number down a lot.
Yes, that is quite the lab floor. Even one per square meter makes a ~780x780 meter space. Very Manhattan Project vibe.
Google tells me Tesla Gigafactory Nevada is about 500k m^2, so about the same :P
Note though that economies of scale and learning curves can make this more economical than it seems. If we assume an experience curve with price per unit going down to 80% each doubling, 600k is 19 doublings, making the units in the last doubling cost just 1.4% of the initial unit cost.
And you don’t need to put all the microscopes in one site. If this were ever to actually happen, presumably it would be an international consortium where there are 10-100 sites in several regions that create technician jobs in those areas (and also reduce the insane demand for 100,000 technicians all in one city).
Also other things might boost efficiency and price.
Most obvious would be nanotechnological systems: likely sooner than people commonly assume, yet might take long enough to arrive to make effect on this project minor if it starts soon. Yet design-ahead for dream equipment might be a sound move.
Advanced robotics and automation is likely major gamechanger. The whole tissue management infrastructure needs to be automated from the start, but there will likely be a need for rapid turnaround lab experimentation too. Those AI agents are not just for doing standard lab tasks but also for designing new environments, tests, and equipment. Whether this can be integrated well in something like the CAIS infrastructure is worth investigating. But rapid AI progress also makes it hard to do plan-ahead.
Biotech is already throwing up lots of amazing tools. Maybe what we should look for is a way of building the ideal model organism—not just with biological barcodes or convenient brainbow coloring, but with useful hooks (in the software sense) for testing and debugging. This might also be where we want to look at counterparts to minimal genomes for minimal nervous systems.
Yet design-ahead for dream equipment might be a sound move.
Thoughts on what that might look like? Are there potentially tractable paths you can imagine people starting work on today?
Imagine a “disassembly chip” that is covered with sensors characterizing a tissue surface, sequences all proteins, carbohydrate chains, and nucleic acids, and sends that back to the scanner main unit. A unit with nanoscale 3D memory storage and near-reversible quantum dot cellular automata processing. You know, the usual. This is not feasible right now, but I think at least the nanocomputers could be designed fairly well for the day we could assemble them (I have more doubts about the chip, since it requires to solve a lot of contingencies… but I know clever engineers). Likely the most important design-ahead pieces may not be superfancy like these, but parts for standard microscopes or infrastructure that are normally finicky, expensive or otherwise troublesome for the project but in principle could be made much better if we only had the right nano or microtech.
So the design-ahead may be all about making careful note of every tool in the system and having people (and AI) look for ways they can be boosted. Essentially, having a proper parts list of the project itself is a powerful design criterion.
What would General Groves do?
Davidad—I recognise we’re coming up on your preferred cutoff time. One other concrete question I’m kind of curious to get your take on, if you’re up for it:
“If you summon your inner General Groves, and you’re given a $1B discretionary budget today… what does your ‘first week in office’ look like? What do you set in motion concretely?” Feel free to splurge a bit on experiments that might or might not be necessary. I’m mostly interested in the exercise of concretely crafting concrete plans.
(also, I guess this might kind of be what you’re doing with ARIA, but for a different plan… and sadly a smaller budget :) )
Yes, a completely different plan, and indeed a smaller budget. In this hypothetical, I’d be looking to launch several FROs, basically, which means recruiting technical visionaries to lead attacks on concrete subproblems:
Experimenting with serial membrane protein barcodes.
Experimenting with parallel membrane protein barcodes.
Build a dedicated brain-imaging synchrotron beamline to begin more frequent experiments with different approaches to stabilizing tissue, pushing the limits on barcoding, and performing R&D on these purported 1-2 OOM speed gains.
Manufacturing organoids that express a diversity of human neural cell types.
Just buy a handful of light-sheet microscopes for doing small-scale experiments on organoids—one for dynamic calcium imaging at cellular resolution, and several more for static expansion microscopy at subcellular resolution.
Recruit for a machine-learning team that wants to tackle the problem of learning the mapping between the membrane-protein-density-annotated-hypergraph that we can get from static imaging data, and the system-of-ordinary-differential-equations that we need to simulate in order to predict dynamic activity data.
Designing robotic assembly lines that manufacture light-sheet microscopes.
Finish the C. elegans upload.
Long-shot about magnetoencephalography.
Long-shot about neural dust communicating with ultrasound.
Some unanswered questions
Me (jacobjacob) and Lisa started the dialogue by brainstorming some questions. We didn’t get around to answering all of them (and neither did we intend to). Below, I’m copying in the ones that didn’t get answered.
What if the proposal succeeds? If the proposal works, what are the…
…limitations? For example, I heard Davidad say before that the resultant upload might mimic patient H.M.: unable to form new memories as a result of the simulation software not having solved synaptic plasticity
…risks? For example, one might be concerned that the tech tree for uploading is shared with that for unaligned neuromorphic AGI. But short timelines have potentially made this argument moot. What other important arguments are there here?
As a reference class for the hundreds of thousands of microscopes needed… what is the world’s current microscope-building capacity? What are relevant reference classes here for scale and complexity—looking, for example, at something like EUV litography machines (of which I think ASML produce ~50/year currently, at like $100M each)?
Some further questions I’d be interested to chat about are:
1. Are there any possible shortcuts to consider? If there are, that seems to make this proposal even more feasible.
1/a. Maybe something like, I can imagine there are large and functional structural similarities across different brain areas. If we can get an AI or other generative system to ‘fill in the gaps’ of more sparse tissue samples, and test whether the reconstructed representation is statistically indistinguishable from say the dynamic data collected in step 4, then we might be able to figure out what density of tissue sampling we need for full predictability. (seems plausible that we don’t need 100% tissue coverage, especially in some areas of the brain?). Note, it also seems plausible to me that one might be missing something important that could show up in a way that wasn’t picked up in the dynamic data, though that seems contingent on the quality of the dynamic data.
1b. Given large amounts of sparsity in neural coding, I wonder if there are some shortcuts around that too. (Granted though that the way the sparsity occurs seems very critical!)
2. Curious to chat a bit more about what the free parameters are in step 5.
3. What might we possibly be missing and is it important? Stuff like extrasynaptic dynamics, glia cells, etc.
3/a. Side note, if we do succeed in capturing dynamic as well as static data, this seems like an incredibly rich data set for basic neuroscience research, which in turn could provide emulation shortcuts. For example, we might be able to more accurately model the role of glia in modulating neural firing, and then be able to simulate more accurately according to whether or not glia are present (and how type of cell, glia cell size, and positioning around the neuron matters, etc).
4. Curious to think more about how to dynamically measure the brain (step 3). Thin living specimens with human genomes and then using the fluorescence paradigm. I’m considering whether there are tradeoffs in only seeing slices at a time where we might be missing data on how the slices might communicate with each other. I wonder if it’d make sense to have multiple sources of dynamic measurements which get combined.. though ofc there are some temporal challenges there, but I imagine that can be sorted out.. like for example using the whole brain ultrasound techniques developed by Sumner’s group. In the talk you mentioned neural dust and communicating out with ultrasound, that seems incredibly exciting. I know UC Berkeley and other Unis were working on this somewhat, though I’m currently unsure what the main blockers here are.
Appendix: the proposal
Here’s a screenshot of notes from Davidad’s talk.
- Shallow review of live agendas in alignment & safety by technicalities (Nov 27, 2023, 11:10 AM; 348 points)
- The Sorry State of AI X-Risk Advocacy, and Thoughts on Doing Better by Thane Ruthenis (Feb 21, 2025, 8:15 PM; 148 points)
- 8 examples informing my pessimism on uploading without reverse engineering by Steven Byrnes (Nov 3, 2023, 8:03 PM; 117 points)
- AI #37: Moving Too Fast by Zvi (Nov 9, 2023, 5:50 PM; 53 points)
- Exploring Whole Brain Emulation by PeterMcCluskey (Apr 6, 2024, 2:38 AM; 13 points)
- Will Aldred's comment on MIRI 2024 Mission and Strategy Update by Malo (EA Forum; Jan 5, 2024, 3:49 PM; 11 points)
- Steven Byrnes's comment on 8 examples informing my pessimism on uploading without reverse engineering by Steven Byrnes (Nov 4, 2023, 12:30 AM; 11 points)
- kman's comment on Significantly Enhancing Adult Intelligence With Gene Editing May Be Possible by GeneSmith (Dec 13, 2023, 7:16 PM; 9 points)
- jacob_cannell's comment on Significantly Enhancing Adult Intelligence With Gene Editing May Be Possible by GeneSmith (Dec 13, 2023, 8:50 PM; 6 points)
- Garrett Baker's comment on D0TheMath’s Shortform by Garrett Baker (Nov 3, 2023, 10:35 PM; 4 points)
- MinusGix's comment on What are the actual arguments in favor of computationalism as a theory of identity? by sunwillrise (Jul 19, 2024, 3:27 AM; 1 point)
I don’t understand what’s being claimed here, and feel the urge to get off the boat at this point without knowing more. Most stuff we care about isn’t about 3-second reactions, but about >5 minute reactions. Those require thinking, and maybe require non-electrical changes—synaptic plasticity, as you mention. If they do require non-electrical changes, then this reasoning doesn’t go through, right? If we make a thing that simulates the electrical circuitry but doesn’t simulate synaptic plasticity, we’d expect to get… I don’t know, maybe a thing that can perform tasks that are already “compiled into low-level code”, so to speak, but not tasks that require thinking? Is the claim that thinking doesn’t require such changes, or that some thinking doesn’t require such changes, and that subset of thinking is enough for greatly decreasing X-risk?
Seconded! I too am confused and skeptical about this part.
Humans can do lots of cool things without editing the synapses in their brain. Like if I say: “imagine an upside-down purple tree, and now answer the following questions about it…”. You’ve never thought about upside-down purple trees in your entire life, and yet your brain can give an immediate snap answer to those questions, by flexibly combining ingredients that are already stored in it.
…And that’s roughly how I think about GPT-4’s capabilities. GPT-4 can also do those kinds of cool things. Indeed, in my opinion, GPT-4 can do those kinds of things comparably well to a human. And GPT-4 already exists and is safe. So that’s not what we need.
By contrast, when I think about what humans can do that GPT-4 can’t do, I think of things that unfold over the course of minutes and hours and days and weeks, and centrally involve permanently editing brain synapses. (See also: “AGI is about not knowing how to do something, and then being able to figure it out.”)
Hm, here’s a test case:
GPT4 can’t solve IMO problems. Now take an IMO gold medalist about to walk into their exam, and upload them at that state into an Em without synaptic plasticity. Would the resulting upload would still be able to solve the exam at a similar level as the full human?
I don’t have a strong belief, but my intuition is that they would. I recall once chatting to @Neel Nanda about how he solved problems (as he is in fact an IMO gold winner), and recall him describing something that to me sounded like “introspecting really hard and having the answers just suddenly ‘appear’...” (though hopefully he can correct that butchered impression)
Do you think such a student Em would or would not perform similarly well in the exam?
I don’t have a strong opinion one way or the other on the Em here.
In terms of what I wrote above (“when I think about what humans can do that GPT-4 can’t do, I think of things that unfold over the course of minutes and hours and days and weeks, and centrally involve permanently editing brain synapses … being able to figure things out”), I would say that human-unique “figuring things out” process happened substantially during the weeks and months of study and practice, before the human stepped into the exam room, wherein they got really good at solving IMO problems. And hmm, probably also some “figuring things out” happens in the exam room, but I’m not sure how much, and at least possibly so little that they could get a decent score without forming new long-term memories and then building on them.
I don’t think Ems are good for much if they can’t figure things out and get good at new domains—domains that they didn’t know about before uploading—over the course of weeks and months, the way humans can. Like, you could have an army of such mediocre Ems monitor the internet, or whatever, but GPT-4 can do that too. If there’s an Em Ed Witten without the ability to grow intellectually, and build new knowledge on top of new knowledge, then this Em would still be much much better at string theory than GPT-4 is…but so what? It wouldn’t be able to write groundbreaking new string theory papers the way real Ed Witten can.
I have said many times that uploads created by any process I know of so far would probably be unable to learn or form memories. (I think it didn’t come up in this particular dialogue, but in the unanswered questions section Jacob mentions having heard me say it in the past.)
Eliezer has also said that makes it useless in terms of decreasing x-risk. I don’t have a strong inside view on this question one way or the other. I do think if Factored Cognition is true then “that subset of thinking is enough,” but I have a lot of uncertainty about whether Factored Cognition is true.
Anyway, even if that subset of thinking is enough, and even if we could simulate all the true mechanisms of plasticity, then I still don’t think this saves the world, personally, which is part of why I am not in fact pursuing uploading these days.
That’s a very interesting point, ‘synaptic plasticity’ is probably a critical difference. At least the recent results in LLMs suggest.
The author not considering, or even mentioning, it also suggests way more work and thought needs to be put into this.
Hello folks! I’m the person who presented the ‘expansion x-ray microtomography’ approach at the Oxford workshop. I wanted to mention that since that presentation, I’ve done a lot of additional research into what would be needed to facilitate 1 year imaging of human brain via synchrotron + ExM. I now have estimates for 11-fold expansion to 27 nm voxel size, which is much closer to what is needed for traceability (further expansion could also help if needed). You can find my updated proposal here: https://logancollinsblog.com/2023/02/22/feasibility-of-mapping-the-human-brain-with-expansion-x-ray-microscopy/
In addition, I am currently compiling improved estimates for the speed of other imaging modalities for comparison. I’ve done a deep dive into the literature and identified the most competitive approaches now available for electron microscopy (EM) and expansion light-sheet fluorescence microscopy (ExLSFM). I expect to make these numbers available in the near future, but I want to go through and revise my analysis paper before posting it. If you are interested in learning more early, feel free to reach out to me!
I don’t think that faster alignment researchers get you to victory, but uploading should also allow for upgrading and while that part is not trivial I expect it to work.
I’d be quite interested in elaboration on getting faster alignment researchers not being alignment-hard — it currently seems likely to me that a research community of unupgraded alignment researchers with a hundred years is capable of solving alignment (conditional on alignment being solvable). (And having faster general researchers, a goal that seems roughly equivalent, is surely alignment-hard (again, conditional on alignment being solvable), because we can then get the researchers to quickly do whatever it is that we could do — e.g., upgrading?)
I currently guess that a research community of non-upgraded alignment researchers with a hundred years to work, picks out a plausible-sounding non-solution and kills everyone at the end of the hundred years.
I’m highly confused about what is meant here. Is this supposed to be about the current distribution of alignment researchers? Is this also supposed to hold for e.g. the top 10 percentile of alignment researchers, e.g. on safety mindset? What about many uploads/copies of Eliezer only?
I’m confused by this statement. Are you assuming that AGI will definitely be built after the research time is over, using the most-plausible-sounding solution?
Or do you believe that you understand NOW that a wide variety of approaches to alignment, including most of those that can be thought of by a community of non-upgraded alignment researchers (CNUAR) in a hundred years, will kill everyone and that in a hundred years the CNUAR will not understand this?
If so, is this because you think you personally know better or do you predict the CNUAR will predictably update in the wrong direction? Would it matter if you got to choose the composition of the CNUAR?
Thanks for making this dialogue! I’ve been interested in the science of uploading for awhile, and I was quite excited about the various C. elegans projects when they started.
I currently feel pretty skeptical, though, that we understand enough about the brain to know which details will end up being relevant to the high-level functions we ultimately care about. I.e., without a theory telling us things like “yeah, you can conflate NMDA receptors with AMPA, that doesn’t affect the train of thought” or whatever, I don’t know how one decides what details are and aren’t necessary to create an upload.
You mention that we can basically ignore everything that isn’t related to synapses or electricity (i.e., membrane dynamics), because chemical diffusion is too long to account for the speed of cognitive reaction times. But as Tsvi pointed out, many of the things we care about occur on longer timescales. Like, learning often occurs over hours, and is sometimes not stored in synapses or membranes—e.g., in C. elegans some of the learning dynamics unfold in the protein circuits within individual neurons (not in the connections between them).[1] Perhaps this is a strange artifact of C. elegans, but at the very least it seems like a warning flag to me; it’s possible to skip over low-level details which seem like they shouldn’t matter, but end up being pretty important for cognition.
That’s just one small example, but there are many possibly relevant details in a brain… Does the exact placement of synapses matter? Do receptor subtypes matter? Do receptor kinematics matter, e.g., does it matter that NMDA is a coincidence detector? Do oscillations matter? Dendritic computation? Does it matter that the Hodgkin-Huxley model assumes a uniform distribution of ion channels? I don’t know! There are probably loads of things that you can abstract away, or conflate, or not even measure. But how can you be sure which ones are safe to ignore in advance?
This doesn’t make me bearish on uploading in general, but it does make me skeptical of plans which don’t start by establishing a proof of concept. E.g., if it were me, I’d finish the C. elegans simulation first, before moving onto to larger brains. Both because it seems important to establish that the details that you’re uploading in fact map onto the high-level behaviors that we ultimately care about, and because I suspect that you’d sort out many of the kinks in this pipeline earlier on in the project.
“The temperature minimum is reset by adjustments to the neuron’s internal signaling; this requires protein synthesis and takes several hours” and “Again, reprogramming a signaling pathway within a neuron allows experience to change the balance between attraction and repulsion.” Principles of Neural Design, page 32, under the section “Associative learning and memory.” (As far as I understand, these internal protein circuits are separate from the transmembrane proteins).
Are y’all familiar with what Openwater has been up to? my hunch is that it wouldn’t give high enough resolution for the things intended here, but I figured it was worth mentioning: it works by doing holographic descattering in infrared through the skull, and apparently has MRI-level resolution. I have wondered if combining it with AI is possible, or if they’re already relying on AI to get it to work at all. The summary that first introduced me to it was their ted talk—though there are also other discussions with them on youtube. I didn’t find a text document which goes into a ton of detail about their tech but somewhat redundant with the ted talk might be one of the things on their “in the press” list, eg this article.
I carefully read all the openwater information and patents during a brief period where I was doing work in a similar technical area (brain imaging via simultaneous use of ultrasound + infrared). …But that was like 2017-2018, and I don’t remember all the details, and anyway they may well have pivoted since then. But anyway, people can hit me up if they’re trying to figure out what exactly Openwater is hinting at in their vague press-friendly descriptions, I’m happy to chat and share hot-takes / help brainstorm.
I don’t think they’re aspiring to measure anything that you can’t already measure with MRI, or else they would have bragged about it on their product page or elsewhere, right? Of course, MRI is super expensive and inconvenient, so what they’re doing is still potentially cool & exciting, even if it doesn’t enable new measurements that were previously impossible at any price. But in the context of this post … if I imagine cheap and ubiquitous fMRI (or EEG, ultrasound, etc.), it doesn’t seem relevant, i.e. I don’t think it would make uploading any easier, at least the way I see things.
This is potentially a naive question, but how well would the imagining deal with missing data? Say that 1% (or whatever the base rate is) of tissue samples would be destroyed during slicing or expansion—would we be able to interpolate those missing pieces somehow? Do we know any bounds on the error would that introduce in the dynamics later?
We know it depends on damage repartition. Losing1% of your neurons is enough to destruct your thalamus, which looks like your brain is dead. But you can also lose much more without noticing anything, if the damage is sparse enough.
Short answer: no.
Even assuming that you can scan whatever’s important[1], you’re still unlikely to have remotely the computing power to actually run an upload, let alone a lot of uploads, faster than “real time”… unless you can figure out how to optimize them in a bunch of ways. It’s not obvious what you can leave out.
It’s especially not obvious what you can leave out if you want the strong AGI that you’re thereby creating to be “aligned”. It’s not even clear that you can rely on the source humans to be “aligned”, let alone rely on imperfect emulations.
I don’t think you’re going to get a lot of volunteers for destructive uploading (or actually even for nondestructive uploading). Especially not if the upload is going to be run with limited fidelity. Anybody who does volunteer is probably deeply atypical and potentially a dangerous fanatic. And I think you can assume that any involuntary upload will be “unaligned” by default.
Even assuming you could get good, well-meaning images and run them in an appropriate way[2] , 2040 is probably not soon enough. Not if the kind of computing power and/or understanding of brains that you’d need to do that has actually become available by then. By the time you get your uploads running, something else will probably have used those same capabilities to outpace the uploads.
Regardless of when you got them running, there’s not even much reason to have a very strong belief that more or faster “alignment researchers” would to be all that useful or influential, even if they were really trying to be. It seems at least as plausible to me that you’d run them faster and they’d either come up with nothing useful, or with potentially useful stuff that then gets ignored.
We already have a safety strategy that would be 100 percent effective if enacted: don’t build strong AGI. The problem is that it’s definitely not going to be enacted, or at least not universally. So why would you expect anything these uploads came up with to be enacted universally? At best you might be able to try for a “pivotal event” based on their output… which I emphasize is AGI output. Are you willing to accept that as the default plan?
… and accelerating the timeline for MMAcevedo scenarios does not sound like a great or safe idea.
… which you probably will not get close to doing by 2040, at least not without the assistance of AI entities powerful enough to render the whole strategy moot.
… and also assuming you’re in a world where the scenarios you’re trying to guard against can come true at all… which is a big assumption
Seems falsified by the existence of astronauts?
I, for one, would be more willing to risk my life to go to cyberspace than to go to the moon.
Separately, I’m kind of awed by the idea of an “uploadonaut”: the best and brightest of this young civilisation, undergoing extensive mental and research training to have their minds able to deal with what they might experience post upload, and then courageously setting out on a dangerous mission of crucial importance for humanity.
(I tried generating some Dall-E 1960′s style NASA recruitment posters for this, but they didn’t come out great. Might try more later)
Another big source of potential volunteers: People who are going to be dead soon anyway. I’d probably volunteer if I knew that I’m dying from cancer in a few weeks anyway.
I don’t think they’re comparable at all.
Space flight doesn’t involve a 100 percent chance of physical death, with an uncertain “resurrection”, with a certainly altered and probably degraded mind, in a probably deeply impoverished sensory environment. If you survive space flight, you get to go home afterwards. And if you die, you die quickly and completely.
Still, I didn’t say you’d get no volunteers. I said you’d get atypical ones and possible fanatics. And, since you have a an actual use for the uploads, you have to take your volunteers from the pool of people you think might actually be able to contribute. That’s seems like an uncomfortably narrow selection.
I think historically folks have gone to war or on other kinds of missions that had death rates of like, at least, 50%. And folks, I dunno, climb Mount Everest, or figured out how to fly planes before they could figure out how to make them safe.
Some of them were for sure fanatics or lunatics. But I guess I also think there’s just great, sane, and in many ways whole, people, who care about things greater than their own personal life and death, and are psychologically consituted to be willing to pursue those greater things.
See the 31 climbers in a row who died scaling Nanga Parbat before the 32nd was successful.
Noting that I gave this a weak downvote as I found this comment to be stating many strong claims but without correspondingly strong (or sometimes not really any) arguments. I am still interested in the reasons you believe these things though (for example, like a fermi on inferece cost at runtime).
OK… although I notice that everybody in the initial post is just assuming you could run the uploads without providing any arguments.
Human brains have probably more than 1000 times as many synapses as current LLMs have weights. All the values describing the synapse behavior have to be resident in some kind of memory with a whole lot of bandwidth to the processing elements. LLMs already don’t fit on single GPUs.
Unlike transformers, brains don’t pass nice compact contexts from layer to layer, so splitting them across multiple GPU-like devices is going to slow you way down because you have to shuttle such big vectors between them… assuming you can even vectorize most of it at all given the timing and whatnot, and don’t have to resort to much discrete message passing.
It’s not even clear that you can reduce a biological synapse to a single weight; in fact you probably can’t. For one thing, brains don’t run “inference” in the way that artificial neural networks do. They run forward “inference-like” things, and at the same time do continuous learning based on feedback systems that I don’t think are well understood… but definitely are not back propagation. It’s not plausible that a lot of relatively short term tasks aren’t dependent on that, so you’re probably going to have to do something more like continuously running training than like continuously running inference.
There are definitely also things going on in there that depend on the relative timings of cascades of firing through different paths. There are also chemicals sloshing around that affect the ensemble behavior of whole regions on the scale of seconds to minutes. I don’t know about in brains, but I do know that there exist biological synapses that aren’t just on or off, either.
You can try to do dedicated hardware, and colocate the “weights” with the computation, but then you run into the problem that biological synapses aren’t uniform. Brains actually do have built-in hardware architectures, and I don’t believe those can be replicated efficiently with arrays of uniform elements of any kind… at least not unless you make the elements big enough and programmable enough that your on-die density goes to pot. If you use any hardwired heterogeneity and you get it wrong, you have to spin the hardware design, which is Not Cheap (TM). You also lose density because you have to replicate relatively large computing elements instead of only replicating relatively dense memory elements. You do get a very nice speed boost on-die, but I at a wild guess I’d say that’s probably a wash with the increased need for off-die communication because of the low density.
If you want to keep your upload sane, or be able to communicate with it, you’re also going to have to give it some kind of illusion of a body and some kind of illusion of a comprehensible and stimulating environment. That means simulating an unknown but probably large amount of non-brain biology (which isn’t necessarily identical between individuals), plus a not-inconsiderable amount of outside-world physics.
So take a GPT-4 level LLM as a baseline. Assume you want to speed up your upload to be able to fast-talk about as fast as the LLM can now, so that’s a wash. Now multiply by 1000 for the raw synapse count, by say 2 for the synapse diversity, by 5? for the continuous learning, by 2 for the extra synapse complexity, and by conservatively 10 for the hardware bandwidth bottlenecks. Add another 50 percent for the body, environment, etc.
So running your upload needs 300,000 times the processing power you need to run GPT-4. Which I suspect is usually run on quad A100s (at maybe $100,000 per “inference machine”).
You can’t just spend 30 billion dollars and shove 1,200,000 A100s into a chassis; the power, cooling, and interconnect won’t scale (nor is there fab capacity to make them). If you packed them into a sphere at say 500 per cubic meter (which allows essentially zero space for cooling or interconnects, both of which get big fast), the sphere would be about 16 meters across and dissipate 300MW (with a speed of light delay from one side to the other of 50ns).
Improved chips help, but won’t save you. Moore’s law in “area form” is dead and continues to get deader. If you somehow restarted Moore’s law in its original, long-since-diverged-from form, and shrank at 1.5x in area per year for the next 17 years, you’d have transistors ten times smaller than atoms (and your power density would be, I don’t know, 100 time as high, leading to melted chips). And once you go off-die, you’re still using macrosopic wires or fibers for interconnect. Those aren’t shrinking… and I’m not sure the dies can get a lot bigger.
Switching to a completely different architecture the way I mentioned above might get back 10X or so, but doesn’t help with anything else as long as you’re building your system out of a fundamentally planar array of transistors. So you still have a 240 cubic meter, 30MW, order-of-3-billon-dollar machine, and if you get the topology wrong on the first try you get to throw it away and replace it. For one upload. That’s not very competitive with just putting 10 or even 100 people in an office.
Basically, to be able to use a bunch of uploads, you need to throw away all current computing technology and replace it with some kind of much more element-dense, much more interconnect-dense, and much less power-dense computing substrate. Something more brain-like, with a 3D structure. People have been trying to do that for decades and haven’t gotten anywhere; I don’t think it’s going to be manufactured in bulk by 2040.
… or you can try to trim the uploads themselves down by factors that end with multiple zeroes, without damaging them into uselessness. That strikes me as harder than doing the scanning… and it also strikes me as something you can’t make much progress on until you have mostly finished solving the scanning problem.
It’s not that you can’t get some kind of intelligence in realistic hardware. You might even be able to get something much smarter than a human. But you’re specifically asking to run a human upload, and that doesn’t look feasible.
Little nitpicks:
“Human brains have probably more than 1000 times as many synapses as current LLMs have weights.” → Can you elaborate? I thought the ratio was more like 100-200. (180-320T ÷ 1.7T)
“If you want to keep your upload sane, or be able to communicate with it, you’re also going to have to give it some kind of illusion of a body and some kind of illusion of a comprehensible and stimulating environment.” Seems like an overstatement. Humans can get injuries where they can’t move around or feel almost any of their body, and they sure aren’t happy about it, but they are neither insane nor unable to communicate.
My much bigger disagreement is a kind of philosophical one. In my mind, I’m thinking of the reverse-engineering route, so I think:
A = [the particular kinds of mathematical operations that support the learning algorithms (and other algorithms) that power human intelligence]
B = [the particular kinds of affordances enabled by biological neurons, glial cells, etc.]
C = [the particular kinds of affordances enabled by CPUs and GPUs]
You’re thinking of a A→B→C path, whereas I’m thinking that evolution did the A→B path and separately we would do the A→C path.
I think there’s a massive efficiency hit from the fact that GPUs and CPUs are a poor fit to many useful mathematical operations. But I also think there’s a massive efficiency hit from the fact that biological neurons are a poor fit to many useful mathematical operations.
So whereas you’re imagining brain neurons doing the basic useful operations, instead I’m imagining that the brain has lots of little machines involving collections of maybe dozens of neurons and hundreds of synapses assembled into a jerry-rigged contraption that does a single basic useful mathematical operation in an incredibly inefficient way, just because that particular operation happens to be the kind of thing that an individual biological neuron can’t do.
On the main point, I don’t think you can make those optimizations safely unless you really understand a huge amount of detail about what’s going on. Just being able to scan brains doesn’t give you any understanding, but at the same time it’s probably a prerequisite to getting a complete understanding. So you have to do the two relatively serially.
You might need help from superhuman AGI to even figure it out, and you might even have to be superhuman AGI to understand the result. Even if you don’t, it’s going to take you a long time, and the tests you’ll need to do if you want to “optimize stuff out” aren’t exactly risk free.
Basically, the more you deviate from just emulating the synapses you’ve found[1], and the more simplifications you let yourself make, the less it’s like an upload and the more it’s like a biology-inspired nonhuman AGI.
Also, I’m not so sure I see a reason to believe that those multicellular gadgets actually exist, except in the same way that you can find little motifs and subsystems that emerge, and even repeat, in plain old neural networks. If there are a vast number of them and they’re hard-coded, then you have to ask where. Your whole genome is only what, 4GB? Most of it used for other stuff. And it seems as though it’s a lot easier to from a developmental point of view to code for minor variations on “build this 1000 gross functional areas, and within them more or less just have every cell send out dendrites all over the place and learn which connections work”, than for “put a this machine here and a that machine there within this functional area”.
I’m sorry; I was just plain off by a factor of 10 because apparently I can’t do even approximate division right.
A fair point up, with a few limitations. Not a lot of people are completely locked in with no high bandwidth sensory experience, and I don’t think anybody’s quite sure what’s going on with the people who are. Vision and/or hearing are already going to be pretty hard to provide. But maybe not as hard as I’m making them out to be, if you’re willing to trace the connections all the way back to the sensory cells. Maybe you do just have to do the head. I am not gonna volunteer, though.
In the end, I’m still not buying that uploads have enough of a chance of being practical to run in a pre-FOOM timeframe to be worth spending time on, as well as being pretty pessimistic about anything produced by any number of uploaded-or-not “alignment researchers” actually having much of a real impact on outcomes anyway. And I’m still very worried about a bunch of issues about ethics and values of all concerned.
… and all of that’s assuming you could get the enormous resources to even try any of it.
By the way, I would have responded to these sooner, but apparently my algorithm for detecting them has bugs...
… which may already be really hard to do correctly…
I have an important appointment this weekend that will take up most of my time, but hope to come back to this after that, but wanted to quickly note:
Why?
Last time I looked into this 6 years ago seemed like an open question and it could plausibly be backprop or at least close enough: https://www.lesswrong.com/posts/QWyYcjrXASQuRHqC5/brains-and-backprop-a-key-timeline-crux
3yrs ago Daniel Kokotajlo shared some further updates in that direction: https://www.lesswrong.com/posts/QWyYcjrXASQuRHqC5/brains-and-backprop-a-key-timeline-crux?commentId=RvZAPmy6KStmzidPF
It’s possible that you (jacobjacob) and jbash are actually in agreement that (part of) the brain does something that is not literally backprop but “relevantly similar” to backprop—but you’re emphasizing the “relevantly similar” part and jbash is emphasizing the “not literally” part?
I think that’s likely correct. What I mean is that it’s not running all the way to the end of a network, computing a loss function at the end of a well defined inference cycle, computing a bunch of derivatives, etc… and also not doing anything like any of that mid-cycle. If you’re willing to accept a large class of feedback systems as “essentially back propagation”, then it depends on what’s in your class. And I surely don’t know what it’s actually doing.
When I think about uploading as an answer to AI, I don’t think of it as speeding up alignment research necessarily, but rather just outpacing AI. You won’t get crushed by an unaligned AI if you’re smarter and faster than it is, with the same kind of access to digital resources.
(I’ve only skimmed/talked to Claude about this post, so apologies if this was already discussed/addressed; also: cross-posted from some Slack/Signal groups on WBE and AI safety.)
Computational requirements for various [especially hi-fi] WBE simulation levels might make applicability to automated safety research mostly useless (even supposing the rest of the technical problems were solved, e.g. scanning, synapse tracing, etc.). E.g. looking at table 9 from page 80 of the WBE roadmap and assuming roughly 10^22 FLOP total computational power available worldwide (see e.g. https://wiki.aiimpacts.org/ai_timelines/hardware_and_ai_timelines/computing_capacity_of_all_gpus_and_tpus): if a spiking neural network simulation is required, this would only allow for < 10,000 automated (WBE) safety researchers; not bad, but not a huge gain in numbers either (compared to current numbers of safety researchers). And it gets (much) worse with more detailed levels of simulation: the next level (electrophysiology) would allow for <=1 WBE.
So I think this weakens the appeal of hi-fi WBEs (for which I don’t think the arguments were a slamdunk even to begin with) even more.
The LessWrong Review runs every year to select the posts that have most stood the test of time. This post is not yet eligible for review, but will be at the end of 2024. The top fifty or so posts are featured prominently on the site throughout the year.
Hopefully, the review is better than karma at judging enduring value. If we have accurate prediction markets on the review results, maybe we can have better incentives on LessWrong today. Will this post make the top fifty?