Note: thank you Mark Budde, cofounder and CEO of Plasmidsaurus, and Maria Konovalova, a growth/marketing/talented-person at Plasmidsaurus, for talking to me for this article! Also thank you to Eryney Marrogi, who helped answer some of my dumb questions about plasmids.

Introduction
Here’s some important context for this essay: it really, really sucks to start a company in biology.
Despite billions in funding, the brightest minds the world has to offer, and clear market need, creating an enduring company here feels almost impossible. Some of this has to do with the difficulties of engaging with the world of atoms, some of it has to do with the modern state of enormously expensive clinical trials, and some of it still can be blamed on something else. To some degree, this is an unavoidable facet of this field; working in it means you’re here for the ‘love of the game’ than anything else.
But is it necessarily fair to equate all for-profit life-science endeavors with grueling, decade-long struggles to bypass scientific obstacles? Is there a world in which life-sciences startups can have a more traditional tech culture ethos in how they approach things? Unfortunately, probably not for a startup aiming to do the traditional therapeutics play.
But if we broaden our scope to companies to include in service provider biotechs, I can offer at least one example: Plasmidsaurus.
Plasmidsaurus was started in 2021 and is currently run by Mark Budde. Some historical context: Mark was the founder of a separate, but related company called Primordium Labs, which merged with another separate company called SNPsaurus. They both were largely doing the same thing, so, circa 2022, they agreed to combine underneath the Plasmidsaurus name.
People who currently work on the wet-lab side of biotech have likely not only heard of this company, but are also loyal customers. On the flip side, I would guess that not even computational folks at biotech companies have heard of them, much less anyone in other industries! This is a shame, and something I’ve been wanting to rectify since I first stumbled across this company.
At their start, they really only did one thing: sequence plasmids. We’ll get into what exactly that means later on in the post, but you can think of it as a sort of on-demand quality assurance for certain lab processes. And the way they do that plasmid sequencing isn’t particularly novel. They use a nanopore sequencer (again, more details later on) to perform it, which is a technology that one, someone else developed a decade ago, and two, a tool you can buy directly from the people who made it.
The workflow here is simple: send them 15 bucks, mail them your plasmids, and within a day, you get back some quality checks on your plasmids of interest. They have leaked into sequencing things outside of plasmids alone (microbial and AAV genome sequencing too), but plasmids are their primary claim to fame.
Despite how uninspiring they seem, how much of a moat they seem to lack, and how cheap their services are, Plasmidsaurus is enormously successful.
For one, they are almost entirely bootstrapped. They have never taken venture capital funding, at most participating in a six-week accelerator run by Fifty Years and taking a small seed investment from them 3 years ago — primarily for networking purposes rather than for money. Even for the niche that Plasmidsaurus is in, a largely bootstrapped biotech is basically unheard of! They were revenue positive from day one.
Secondly, despite the bootstrapping, they are consistently growing. They are currently at 40 employees, with 225% growth over the last 2 years and 34% growth over the last 6 months (according to LinkedIn). They currently have nine sequencing labs across the globe and have plans to open up even more.
Thirdly and finally, their customers are incredibly happy with them. This is something you don’t see amongst very many biotech services providers, at least the ones I’m aware of. I know of Twist Biosciences as another one that inspires the same level of enthusiasm, but, besides that, it’s uncommon. Here is a picture of several Reddit comments I found, all singing their praises. And these aren’t cherry-picked; I struggle to find even a single post that says something bad about Plasmidsaurus.
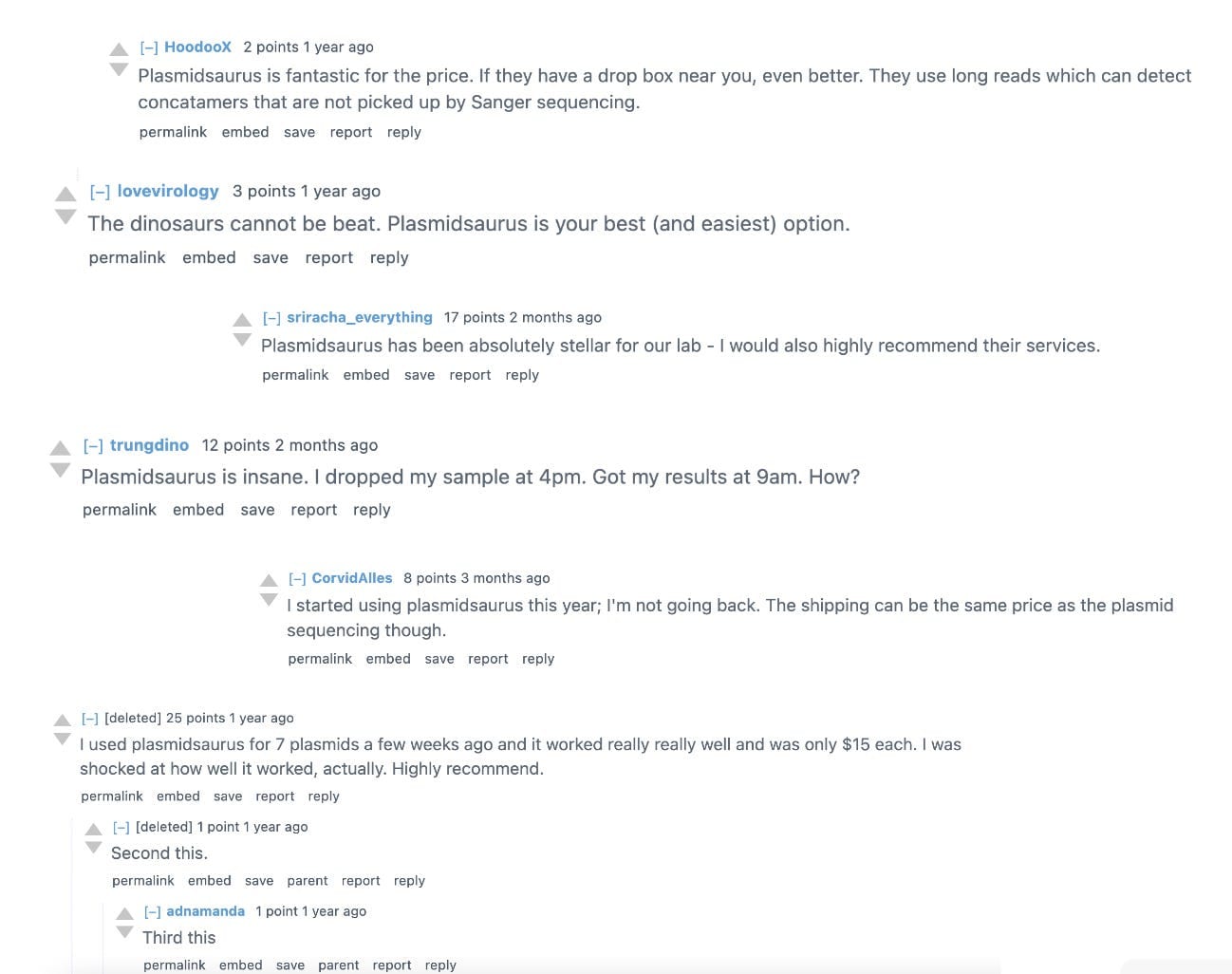
The sum combination of these three characteristics make Plasmidsaurus unique amongst nearly every biotech company I’m aware of. This essay will explain the scientific fundamentals of the company, what niche they occupy, how they managed to grow so much despite their simplicity, and, as always, the risks that the company has.
Background
What is a plasmid?
Plasmids are small, circular pieces of DNA that exist separately from an organism’s main chromosomal DNA. They naturally occur in bacteria, but not in most other domains of life (with some exceptions).
Why do they exist? Answering that question has the same challenges of asking why anything in biology exists, but one possible reason is for their role in horizontal gene transfer. This is the practice of a bacteria sharing potentially useful genes with other neighboring bacteria, and the structure of plasmids lends itself well to being shunted out a bacterium and absorbed by others. Of course, they likely evolved for a plethora of reasons beyond gene transfer alone, but we’ll accept the surface level explanation for now.
Why do people care about plasmids? First, a brief tangent: most DNA you’ll find in a cell will be somewhat of a mess; all sorts of uncharacterized regulatory elements, genes, and the like. Of course, it all makes sense to the cell that holds it, but it is a bit incomprehensible to a human. This is often a barrier to scientists who are interested in modifying behaviors of that cell, such as using it to produce a specific protein. But, as it happens, most cells generally don’t care about where a piece of genetic material came from; even if it’s not part of their own DNA, just floating around! If genetic material shows up somewhere where there are transcription proteins around, cells will happily use it. This is, shockingly, as true for bacterium as it is for mammalian cells, even though mammalian cells don’t naturally have plasmids!
This is the primary application of plasmid: a single piece of genetic material, stripped down to the essentials, and extremely modifiable by a researcher, used for the purpose of modifying cellular behavior. A single plasmid can encode for multiple genes — with regulation logic on top of those genes — and continue to survive in cells even as they divide. Once the plasmid enters a cell nucleus, which is done by placing the plasmid near a cell and applying an electric field to increase the cell membranes permeability (along with other possible techniques), it will be interpreted the same as any other source of genetic material. A scientist needn’t modify the raw DNA of a cell to change how it functions, introducing a plasmid into the cell is often sufficient!
Again, given how useful they are, plasmids are present in nearly every modern biology experiment. Which is why it’s a bit troubling that an immense number of plasmids likely have errors in them.
Errors in plasmids
In a BiorXiv preprint from August 2024 titled ‘Prevalence of errors in lab-made plasmids across the globe’, the authors stumble across a worrying statistic:
We found that approximately 15% of plasmids had significant design errors, and about 35% contained sequence errors in functional regions…in total, we estimate that 45-50% of lab-made plasmids have undetected design and/or sequence errors that could potentially compromise the intended applications. Indeed, we suspect that this figure may underestimate the true scale of quality issues in lab-made plasmids because we had asked our clients to check the designs and sequences of their plasmids before submission to us, and also because they were paying for our services utilizing their plasmids.
The ‘design errors’ here isn’t a massive deal, since those are more along the lines of ‘the person who designed the plasmid messed up’. Plasmids, despite their simplicity, have their own biophysical limitations. And it’s very possible to accidentally create a plasmid that is a-priori known to be bad at its job. This is a problem, but also an avoidable one.
Some examples of (avoidable) design errors
The much more concerning bit are the sequence errors. Those aren’t something you can predict in advance, they just happen accidentally as a result of biophysical instability, accidental contamination, or chemical bad luck. From the authors:
Notably, about 35% of these plasmids (91/259) displayed sequence variations from the senders’ reference (Figure 1B). Among them, we identified 89 point mutations, 35 deletions, and 19 insertions, with some plasmids containing multiple types of error.
Stuff like this can dramatically change the takeaway message from biology studies. After all, single mutations can destabilize an entire protein! It’d be inaccurate to fault plasmid errors as the singular reason for why biology is undergoing a replication crisis — there are far more possible sources — but it certainly doesn’t help.
The value add of Plasmidsaurus
It is within the extreme importance of plasmids, alongside how error prone they are to work with, that Plasmidsaurus found its niche: in ensuring your plasmids actually match up to what you want them to be.
Let’s restate their business model: you send them the plasmid you’ve produced, give them $15, they sequence it, and tell you if it matches what you expect the plasmid to look like — all usually in less than a day. With that, we can go through some immediate questions.
How do they sequence it? As mentioned before, using a nanopore sequencer. Here are some more details on what that exactly is, but all you really need to know is that it is fast, cheap, and a very convenient way to sequence things. The last part is important; it means scientists using a nanopore need to do very little ‘prep’ work for their plasmid to be successfully sequenced. Nanopores can just handle it, whereas other sequencing methods (e.g. Sanger sequencing) require some extra work and have more failure modes.
Did Plasmidsaurus come up with with nanopores? No, the whole concept is the intellectual property of Oxford Nanopore, who independently sells nanopores, Plasmidsaurus is just partnered with them.
Wait…so I can just buy a nanopore? Is it just really expensive? It’s actually really cheap for a sequencing machine. Oxford Nanopore’s MinION device, their most portable sequencer, costs around $2,000 for the starter pack. For reference, the cost of non-nanopore sequencing machines can reach six figures. Fairly, the cost of actually running each nanopore sequencing run can also reach into the hundreds-of-dollars in terms of the materials required, but still, not much for many labs.
Then…can’t I just do what Plasmidsaurus is doing myself? What’s the point of them? This is an incredibly fair question and exemplifies what I find really interesting about Plasmidsaurus.
So, nanopore sequencing has two unique characteristics.
An improvement on the state-of-the-art. Because sequencing is so widespread in biology, anything that can help push sequencing even somewhat further is incredibly useful. Nanopores are simultaneously improvements on cost, convenience, and sequencing length than competing approaches.
Culturally new. Nanopore sequencing is a newcomer to the sequencing scene. Traditional methods like Sanger sequencing or short-read NGS platforms (e.g. Illumina) have been around for decades, while nanopores were commercially introduced in 2014. Because of this, it hasn’t had time to become a standard in-house technique for most labs. While the nanopore platform becomes better year-after-year, currently, most researchers find them to be incredibly hard to use. As an example of their finickiness, they were famously found to work better in a dark room (circa 2022~) for reasons that are still badly understood. Oxford Nanopore, the developer behind Nanopore, is partially to blame; the company is known as a particularly egregious example of ‘incredible technology, terrible market execution’. But it’s also just time, new tools take a while to seep into the normal wet-lab workflow.
If a certain tool is useful, but hard to use, the lack of entrenchment of that tool creates opportunities for specialized service providers. It is upon this hill that Plasmidsaurus has built their business.
Of course, Plasmidsaurus isn’t just a parasite on top of Oxford Nanopore, there is a huge amount of optimization they have internally done with nanopores to ensure high quality data at a low price point. Specifically, a vast network of error-checking software and optimized protocols, none of which is public knowledge. Moreover, they also have an established relationship with Oxford Nanopore, allowing them much deeper insight into new products, chemistries, and tools that the company is developing. And even a chance to influence it! Finally, they also work hard to ensure logistical efficiency; there are hundreds of Plasmidsaurus drop-boxes and nine sequencing labs placed around the world to ensure that sending them a plasmid + getting back the results is as fast and convenient as possible.
But they are nevertheless a strong deviation from typical biotech companies. You could draw an interesting parallel from Plasmidsaurus to Databricks — a now 43-billion dollar company who transformed the use of Apache Spark in the data engineering world.
You could set up and manage your own Spark cluster, dealing with all the intricacies of distributed computing, resource allocation, and optimization. Or you could turn to Databricks, pay a premium, and focus on your data munging code without worrying about the underlying infrastructure. Some might argue that relying on Databricks creates a single point of failure and discourages deep understanding of the technology. But others would contend that Databricks has been instrumental in making Spark accessible to a wider audience, allowing analysts to concentrate on deriving insights rather than managing complex systems.
Plasmidsaurus isn’t dissimilar to companies like Databricks. By leveraging a relatively new sequencing platform, optimizing their processes to offer an incredibly low price point, and ensuring rapid turnaround times (less than a day), they are seamlessly integrating themselves into the workflow of many labs. Much like how Databricks didn’t invent Spark (though, admittedly, one of the founders did so during their PhD), Plasmidsaurus isn’t inventing new sequencing methods, they are instead making already-invented methods much, much easier to work with, and charging a very small premium on top of that. Labs who otherwise would otherwise never do comprehensive plasmid quality checks have now incorporated it into their typical workflow. It’s very much like a traditional software-as-a-service business!
While this sort of mentality is common enough amongst tech-centric companies to the point where it’s considered old-hat, it’s genuinely innovative in the context of biology. Though wet-lab research contains traces of this outsourcing culture — otherwise, service providers wouldn’t exist at all — there is, anecdotally, a stronger hesitation with relying on them. Good service providers are hard to find (and their ‘goodness’ can disappear over time), often unreliable, and can take weeks to get information back from. While labs will rely on these external companies to handle experiments that they are literally incapable of doing with the equipment they have in-house, the preference is to keep things internal.
Plasmidsaurus’ growth depended on changing that culture.
Their growth
Interestingly, Mark noted that nanopore sequencing for plasmids was not on most people’s radar when he first began offering it as a service in 2021. In fact, nobody believed it could be a viable business.
Nanopores were thought of as extremely error prone, something that was impossible to easily address. Moreover, few believed that changes to one area of the plasmid could affect other regions. As in, full plasmid sequencing was overkill, partial sequencing using traditional methods was sufficient. Mark had worked with using nanopores for plasmid sequencing years earlier — back in 2015 when they were first released to application-only users — and had first-hand experience in knowing that both of these opinions were fundamentally wrong. Not only was full plasmid sequencing far more important than most believed, nanopores with the right bioinformatics tooling could achieve it with high accuracy.
At the time, he vaguely thought a business could be made here. Six years later, he decided to do exactly that, initially running the whole operation out of his garage.
Because of peoples doubts and the aforementioned reluctance of biology research to rely on outsourcing, Mark said that the early days of Plasmidsaurus — named Primordium Labs at its founding — was dominated by a Do Things That Don’t Scale mentality. In other words, lots of manual, grassroots user acquisition, hyper-focused on making sure they were happy. Universal amongst tech founders, but rare to find in biology founders, who often assume that the market will come to them if their products are Good Enough.
When the company first began, Mark would reach out to old lab-mates at Caltech (where he did his postdoc) and offered to sequence their plasmids for free. He’d walk on over to nearby industry labs with candy and a sales pitch for why they should use his services. He primarily targeted top, Nobel-prize-winning research groups — as they were typically both open-minded and had money to spend — with the hopes that other labs would follow their lead.
I also talked with Maria Konovalova — who manages growth at Plasmidsaurus — about this further. She emphasized how the playful aspect of Plasmidsaurus marketing was surprisingly fruitful in gaining more attention in communities they care about. For example, Plasmidsaurus made cereal boxes that had sequencing ads on them to hand out at conferences!

Plasmidsaurus has historically done very little ‘traditional’ marketing — no brochures, few cold reach-outs, and (until recently) no sales team at all. Their marketing seems intended to simply pique your curiosity as to who this dinosaur-centric company is. As another example, they released this video of a guy in a dinosaur suit doing BMX stunts while carting around a box of plasmid sequences. All to announce their new lab in London, which further drove down time-to-sequence for companies in the area.
Marketing like this is risky! It’s hard to a-priori tell whether this sort of branding will have unintended second order consequences. It’s the norm for biotech service companies to have a stoic, corporate, and dispassionate vibe about their whole business for a reason; there is always the risk of not being taken seriously. But in Maria’s eyes, the bet on a more human, personal element to their company has paid off.
Of course, marketing should be backed up by utility, especially in hard science fields where money is tight. Plasmidsaurus combined this marketing with an aggressive focus on improving customer satisfaction. There were no back-and-forth emails on experimental specifics of the sequencing, only an online form for a customer to fill out at their convenience. All pricing was upfront and not hiding behind a sales representative. There was a guaranteed turnaround time of <1 day for biotech hubs, 1-2 days in areas where there isn’t a lab nearby, a level of speed assisted by the 500 dropboxes and 9 sequencing labs they have placed around the world. Because of how surprisingly pleasant Plasmidsaurus was to work with compared to almost every biotech service provider, anyone who tried them once was immediately made a loyal customer.
I think their whole growth story is so interesting. Plasmidsaurus is almost doing a form of cultural arbitrage; applying the fast-paced, customer-centric ethos of tech startups to the traditionally slow-moving world of biotech services. In a field where DIY is often seen as a virtue, they are making it actively desirable to outsource certain tasks. All by using the principles that other fields have known about for decades!
And, even in year 3 of their existence, they still are focused on improving. Increasingly, their sequencing labs are a fleet of automated Opentron machines, further driving up the speed of rendered services. They are aggressively investing into exploring sequencing areas outside of pure plasmid sequencing, including sequencing AAV genomes and sequencing microbial cultures — both of which have become solid businesses in of themselves at this point. Finally, most curiously, Mark suggested to me that they hope to eventually make a software-focused play into assisting the analysis of Plasmidsaurus-produced data. The end goal of everything being to make life-scientific research faster, easier, cheaper, and more replicable.
Potential risks
I’ve been effusively positive about Plasmidsaurus in this essay — perhaps annoyingly so — but all biotechs are fundamentally risky endeavors. Even Plasmidsaurus, despite how good of a position they are in today, have their own bets they are taking. I can think of two distinct risks.
Commoditization of nanopore sequencing.
Right now, nanopore sequencing is somewhat of an arcane art. As we’ve discussed, that’s partially what makes Plasmidsaurus so valuable, they have wrapped it up in an easy-to-use service for use in plasmid sequencing. But what happens when nanopore sequencing becomes as straightforward as running a PCR? Oxford Nanopore, the company behind the technology, is slowly improving their documentation of the whole system. If, in a few years, any lab tech can run nanopore sequencing easily, the barrier to in-house sequencing drops dramatically. At that point, most labs might start to question why they’re outsourcing plasmid sequencing at all.
While I view this as the primary risk that Plasmidsaurus faces, it also isn’t too bad. After all, sequencing is just one part of the pipeline that Plasmidsaurus has built. Having a good user experience — something that Plasmidsaurus has nailed — over what seems like a commoditized process can lead to a lot of otherwise unexpected ‘stickiness’ amongst users. But biotech is also a money-constrained business, so services like Plasmidsaurus may be the first to be cut in economic downswings.
Competition from other sequencing companies.
Big players like Illumina or even Oxford Nanopore themselves could start offering specialized plasmid sequencing services. With their vast resources and established infrastructures, they could potentially undercut Plasmidsaurus on price or bundle plasmid sequencing with other services they already provide. Imagine if labs could get their plasmid sequencing done alongside all other genomic services in one fell swoop—it might be tempting for them to switch. Moreover, new startups might emerge, inspired by Plasmidsaurus’s success but aiming to do it better, faster, or cheaper.
But, again, while this is a risk, I view it as relatively minor. The relationships that Plasmidsaurus has built with their customers is going to be hard to replicate. Labs aren’t just buying a sequencing service; they’re buying reliability, speed, and a hassle-free process that integrates smoothly into their workflow, and even if a new company claims to offer that, only Plasmidsaurus has actually proven themselves. That will likely buy them a lot of time, but the onus is on them to continue innovating to keep that relationship.
Overall, Plasmidsaurus is a startup that has done incredible things, and I fully expect them to continue that trend.
Most importantly though, learning about Plasmidsaurus has been a big mental update to my mental conception of ‘all biotech startups need to solve deep scientific problems and acquire tens-of-millions of dollars in venture funding to get anywhere’. Biology is a huge area that goes beyond therapeutics alone; there are incredibly impactful and profitable businesses to be made across this field. And some of them can resemble the ethos of software development far more than the ethos of drug development.
If you work in a lab and want to check that your plasmid is accurate, you should try out Plasmidsaurus!
and
seem to be a bit contradictory?
Yeah I can see that; I guess what I was trying to get across was that Plasimidsaurus did do a lot of cold reachout at the start (and, when they did do it, it was high-effort, thoughtful reachouts that took into account the labs needs), but largely stopped afterwords.