This is an archival link-post for our preprint, which can be found here.

Executive Summary
This post is a heavily slimmed down summary of our main paper, linked above. We have omitted all the technical details here. This post acts as a TL;DR archival link-post to the main paper.
We train Sparse Autoencoders (SAEs) on the class token of a radiology image encoder, on a dataset of chest x-rays. We use the trained SAE, in conjunction with automated interpretability, to generate radiology reports. The final radiology report represents a concatenation of the text descriptions of activate SAE features. We train a diffusion model to allow causal interventions on SAE features. This diffusion model enables us to highlight where in the chest x-ray each sentence in the radiology report comes from by localising changes in the image post-intervention. Our method achieves competitive accuracy in comparison to state of the art medical foundation models while using a fraction of the parameter count and compute costs. To the best of our knowledge, this is the first time SAEs have been used for a non-trivial downstream task—namely to perform multi-modal reasoning on medical images.
Of particular note to the mechanistic interpretability community, we demonstrate that SAEs extract sparse and interpretable features on a small dataset (240,000) of homogenous images (chest x-rays appear very homogenous), and that these features can be accurately labeled by means of automated interpretability to produce pathologically relevant findings.
Motivation
Radiological services are essential to modern clinical practice, with demand rising rapidly. In the UK, the NHS performs over 43 million radiological procedures annually, costing over £2 billion, and demand for scans more than doubled between 2012 and 2019. A significant portion of these costs addresses rising demand through agency, bank, and overtime staff, but a national imaging strategy notes this funding is unsustainable. Consequently, there’s growing interest in (semi)-automating tasks like radiology report generation, augmentation, and summarization to assist clinicians, spurred by advances in multimodal text-vision modelling techniques.
Recent architectures that combine vision encoders with pretrained Large Language Models (LLMs) to create multimodal Vision-Language Models (VLMs) have shown impressive performance in visual and language tasks. VLMs have been applied to healthcare tasks, including radiology report generation, typically by mapping image representations into the LLM’s token embedding space. The LLM is fine-tuned to respond to prompts like ‘
Despite improvements from scaling VLMs, hallucinations and disagreements with domain experts remain common. Hallucinations are unavoidable in LLMs, and whilst this represents a limitation of current VLM systems designed for radiology report generation, there are other important considerations of using such a system for this task. For current state-of-the-art systems, it is necessary to finetune a multi-billion parameter LLM (as well as projector weights) to perform visual instruction tuning, which is computationally intensive and can be prohibitively expensive. Additionally, the generated reports a VLM provides may not be faithful to the underlying computations of the image encoder – we should aim to design a framework that is verifiably faithful to the image model by reverse engineering the computations of the image encoder. This could yield more interpretable results and thus engender more trust in automated radiology reporting systems.
To this end, we introduce ‘SAE-Rad’, a framework which leverages SAEs, to directly decompose image class tokens from a pre-trained radiology image encoder into human-interpretable features.
Radiology Reporting Pipeline
Please see the pre-print on ArXiv for the detailed experimental setup. We trained an SAE with an expansion factor of 64. The SAE used had the architecture of a gated SAE but without normalising the decoder weights. At the end of training, the SAE had an and an explained variance of . Figure 2 displays the radiology reporting pipeline.
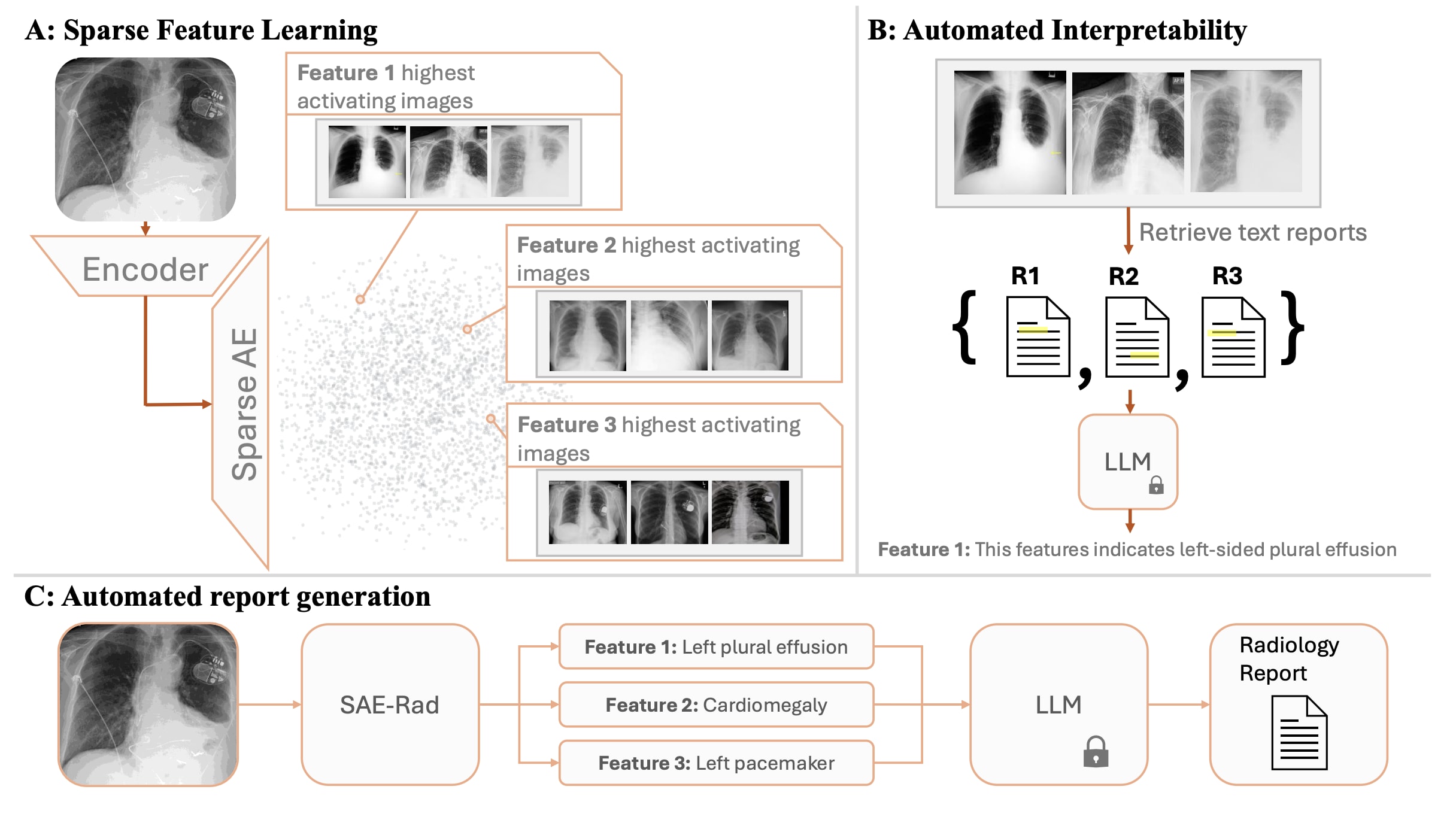
Results
Quantitative Evaluation
For details of the metrics and datasets reported in this section, please refer to the preprint.
We compared SAE-Rad to the current state-of-the-art radiology reporting systems. CheXagent is an instruction-tuned foundation model for CXRs trained on 1.1M scans for question-answering and text-generation tasks. MAIRA-1 &-2 are VLMS based on the LLaVA 1.5 architecture. MAIRA-2 is trained on 510,848 CXRs from four datasets and sets the current state-of-the art for report generation. The MAIRA systems are not publicly available for result replication, and thus we quote their evaluation values directly as our upper-bound. CheXagent is publicly available, and we therefore performed independent replications for this model for a direct comparison.
As Table 1 demonstrates, SAE-Rad underperforms on generic NLG metrics such as BLEU-4. This is expected as we do not try to optimize for any specific ‘writing style’ by fine-tuning an LLM on the reference reports. Conversely, SAE-Rad demonstrates strong performance on radiology-specific metrics which are clinically relevant, outperforming CheXagent by up to 52% in the CheXpert F1 score (macro-averaged F1-14), and achieving 92.1% and 89.9% of the performance of MAIRA-1 and MAIRA-2 on these scores, respectively.
SAE Features
Figure 1 displays some clinically relevant SAE features. In this section we showcase highest activating images for a number of other features, as well as the corresponding feature explanations. We highlight the variety of features captured by SAE-Rad, from instrumentation features to visual features such as radiograph inversion, pathology-related features, and small details such as piercings.

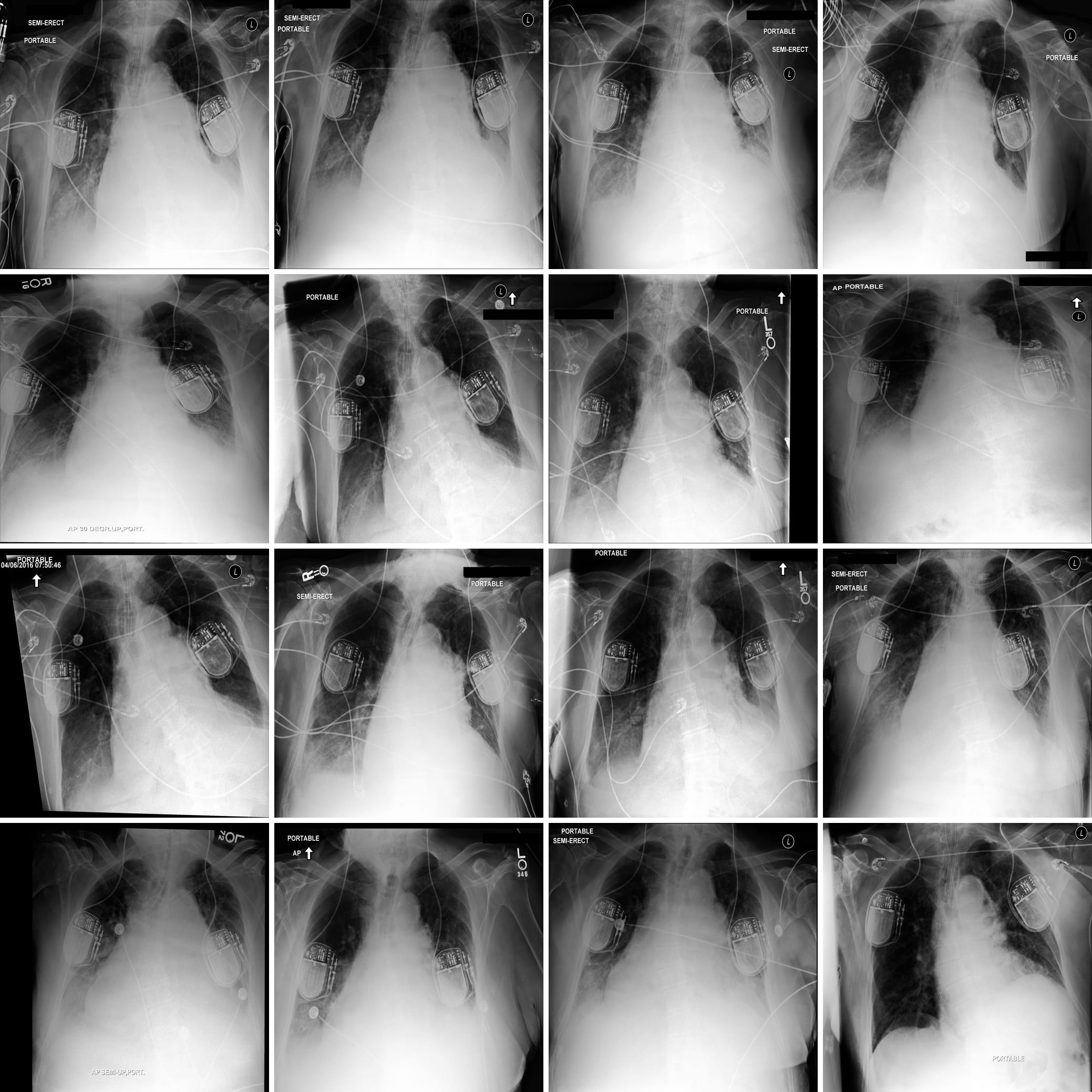
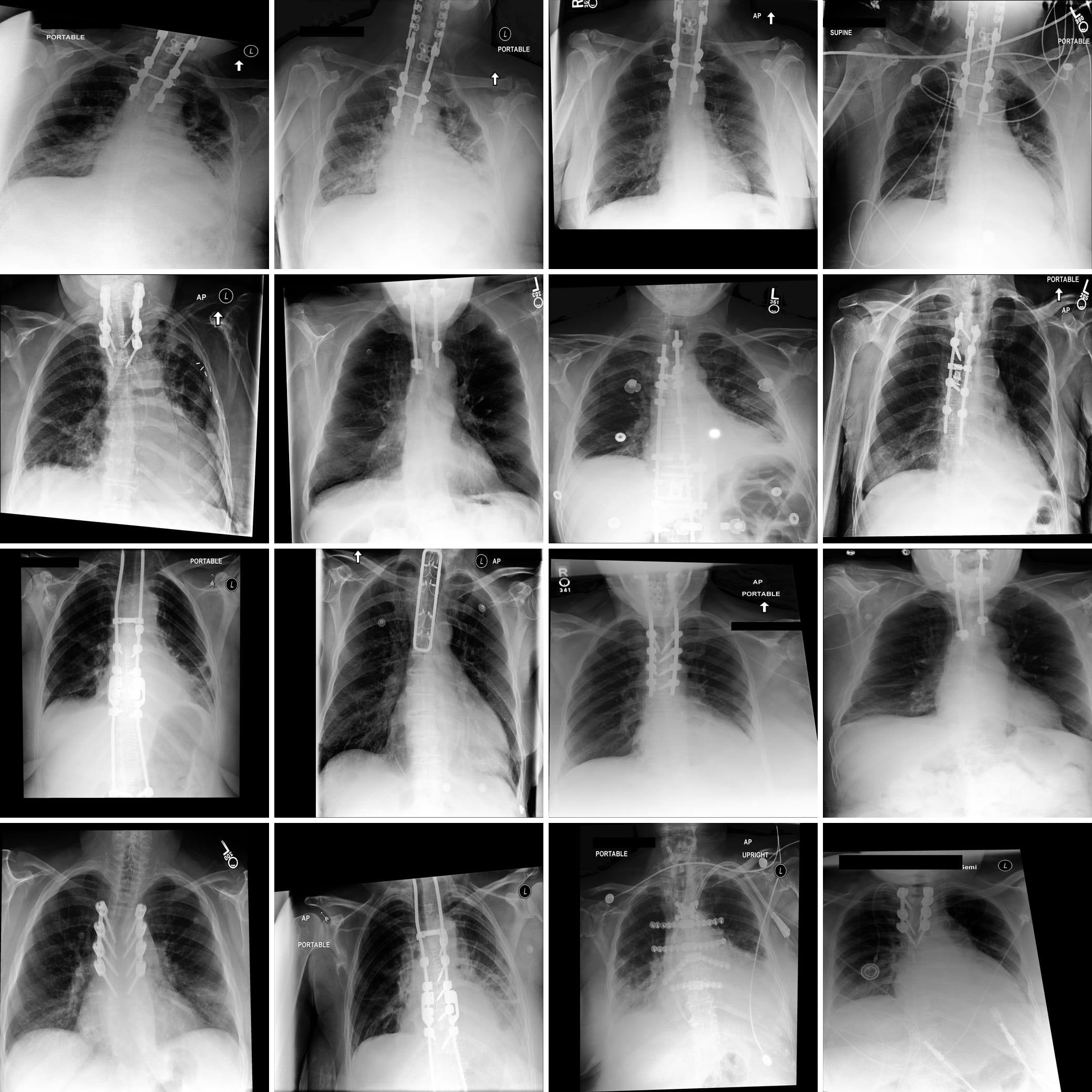
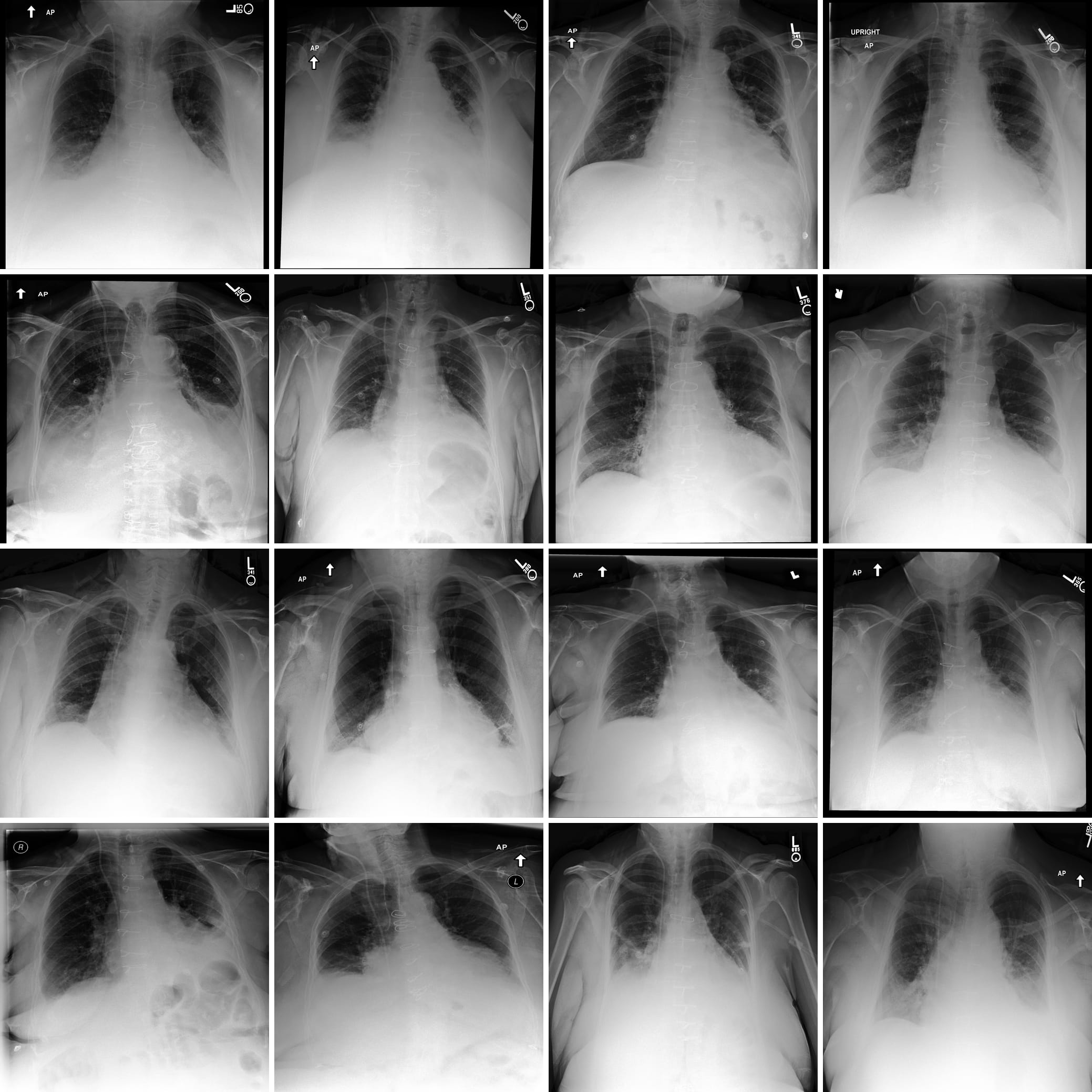
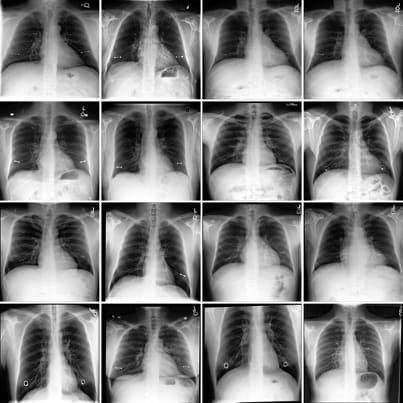
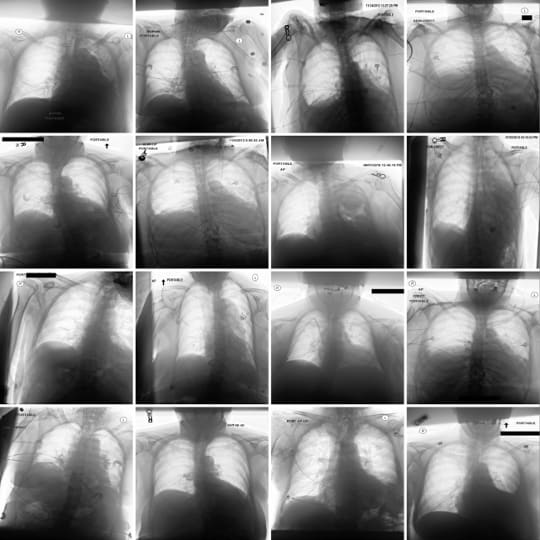

Counterfactual Image Generation
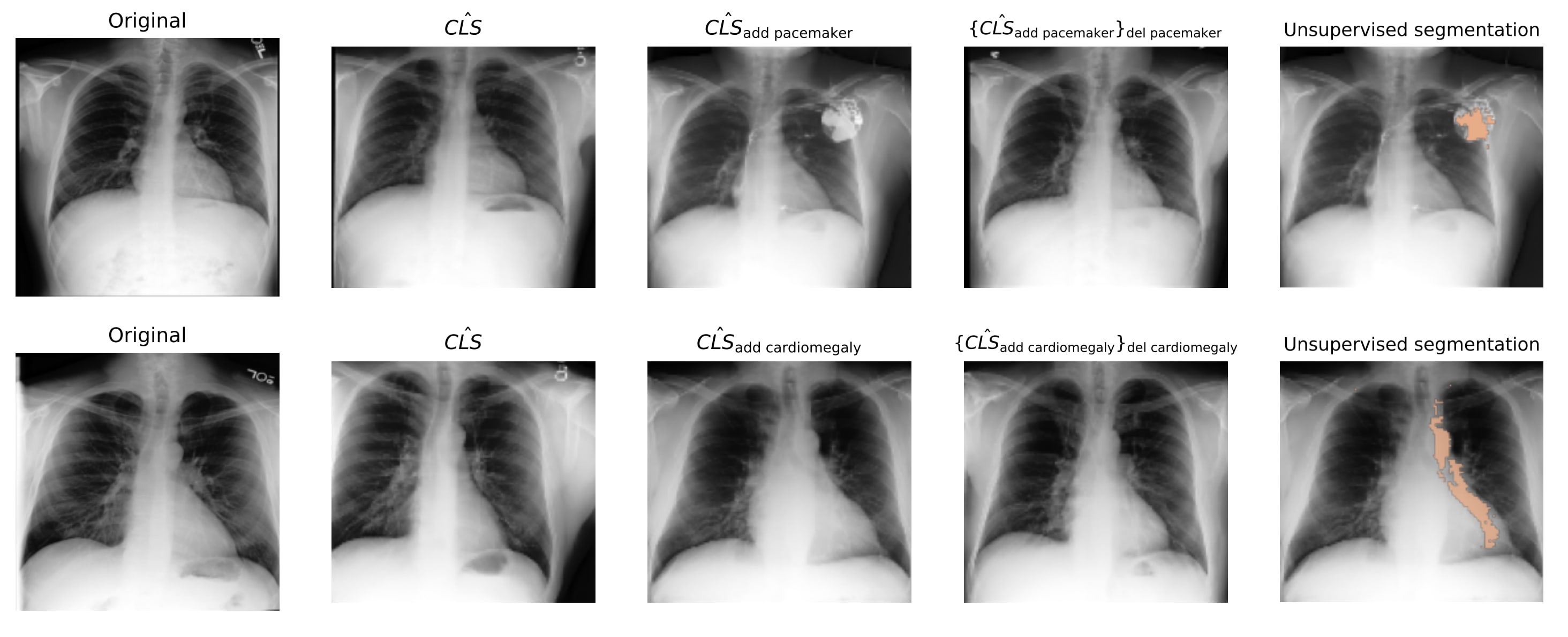
We evaluated the interpretability and validity of our SAE features by intervening on SAE features and then reconstructing the resulting x-rays through a diffusion model. SAE features are interpretable if they correspond to distinct concepts that respond predictably to activation space interventions. We trained a diffusion model conditioned on the class tokens of a radiology image encoder, to reconstruct the radiographs. During inference, we passed a class token through the SAE, intervened on the hidden feature activations, and reconstructed a “counterfactual” class token via the SAE decoder, which conditioned the diffusion model to project interventions into imaging space. We tested whether: 1) interventions alter the reconstructed class token accordingly, 2) changes affect only the targeted feature, and 3) features can be “added” or “removed” by manipulating the same activation. Figure 3 shows the results for two features (cardiomegaly and pacemaker), demonstrating that our interpretations accurately reflect their impact on model behaviour. Figure 3 also illustrates how these methods can be used to ground the radiology report in the chest x-rays through unsupervised segmentation.
Thanks for the great work. I think that multimodal sparse auto encoders is a promising direction. Do you think it is possible / worthwhile to train SAEs on vla models like OpenVLA? I haven’t seen any related work training or interpreting action models using SAE work, and am curious of your thoughts.